Gautschi User Guide
Gautschi is a Community Cluster optimized for communities running traditional, tightly-coupled science and engineering applications.
Link to section 'Overview of Gautschi' of 'Overview of Gautschi' Overview of Gautschi
Gautschi is a Community Cluster optimized for communities running traditional, tightly-coupled science and engineering applications. The Gautschi Community Cluster is equipped with both CPU and GPU compute nodes, each designed for specific computational tasks. The cluster includes Dell CPU compute nodes that feature dual 96-core AMD Epyc "Genoa" processors, providing 192 cores per node and 384 GB of memory. GPU compute nodes come with two 56-core Intel Xeon Platinum 8480+ processors, a total of 1031 GB of CPU memory, and eight NVIDIA H100 GPUs, each boasting 80 GB of memory. All compute nodes have 400 Gbps NDR Infiniband interconnect and service through 2030.
Access to the Gautschi cluster is being offered in units of 48-cores or one quarter share of a CPU node or 5 GPU-year package for GPU nodes. To purchase access to Gautschi today, go to the Cluster Access Purchase page. Please subscribe to our Community Cluster Program Mailing List to stay informed on the latest purchasing developments or contact us via email at rcac-cluster-purchase@lists.purdue.edu if you have any questions.
Link to section 'Gautschi Namesake' of 'Overview of Gautschi' Gautschi Namesake
Gautschi is named in honor of Dr. Walter Gautschi, the Professor in the Department of Computer Science and Mathematics at Purdue. More information about his life and impact on Purdue is available in a Biography of Gautschi.
Link to section 'Gautschi Specifications' of 'Overview of Gautschi' Gautschi Specifications
All Gautschi CPU compute nodes have 192 processing cores, 384 GB of memory, and 400 Gbps NDR Infiniband interconnects. Each Gautschi GPU compute node has eight NVIDIA H100 GPUs with 80 GB of GPU memory, 112 processing cores, 1032 GB of CPU memory, and 400 Gbps NDR Infiniband interconnects.
| Front-Ends | Number of Nodes | Processors per Node | Cores per Node | Memory per Node | Retires in |
|---|---|---|---|---|---|
| 8 | Two AMD EPYC 9654 96-Core Processor “Genoa” CPUs @ 2.4 GHz | 192 | 768 GB | 2030 |
| Sub-Cluster | Number of Nodes | Processors per Node | Cores per Node | Memory per Node | Retires in |
|---|---|---|---|---|---|
| A | 338 | Two AMD EPYC 9654 96-Core Processor “Genoa” CPUs @ 2.4 GHz | 192 | 384 GB | 2030 |
| B | 6 | Two AMD Epyc 9654 96-Core Processor “Genoa” CPUs @ 2.4GHz | 192 | 1.5 TB | 2030 |
| G | 6 | Two AMD EPYC 9554 64-Core Processor “Genoa” CPUs @ 3.1GHz, Two NVIDIA L40S GPUs (48GB) |
128 | 384 GB | 2030 |
| H | 20 | Two Intel Xeon Platinum 8480+ 56-Core CPUs @ 3.8GHz, Eight NVIDIA H100 GPUs (80GB) |
112 | 1 TB | 2030 |
Gautschi nodes run Rocky Linux 9 and use Slurm (Simple Linux Utility for Resource Management) as the batch scheduler for resource and job management. The application of operating system patches occurs as security needs dictate.
On Gautschi, the following set of compiler and message-passing libraries for parallel code are recommended:
- GCC 14.1.0
- OpenMPI

Link to section 'Walter Gautschi' of 'Biography of Gautschi' Walter Gautschi
Professor Gautschi is the Professor Emeritus of Computer Science and Mathematics in the Department of Computer Science at Purdue. Before coming to Purdue, Professor Gautschi did postdoctoral work as a Janggen-Pöhn Research Fellow at the National Institute of Applied Mathematics in Rome and at the Harvard Computation Laboratory. He also held positions at the National Bureau of Standards, the American University, the Oak Ridge National Laboratory, and the University of Tennessee. Since coming to Purdue, he has been a Fulbright Scholar at the Technical University of Munich and has held visiting appointments at the University of Wisconsin, Argonne National Laboratory, the Wright-Patterson Air Force Base, ETH Zurich, the University of Padova, and the University of Basel.
He has been a Fulbright Lecturer, an ACM National Lecturer, and a SIAM Visiting Lecturer. He is, or has been, on the editorial boards of SIAM Journal on Mathematical Analysis, Numerische Mathematik, Calcolo, and Mathematics of Computation, and has served as a special editor for Linear Algebra and Its Applications. From 1984 to 1995, he was the managing editor of Mathematics of Computation and, since 1991, an honorary editor of Numerische Mathematik. In 2001, Professor Gautschi was elected a Corresponding Member of the Bavarian Academy of Sciences and Humanities and, in the same year, a Foreign Member of the Academy of Sciences of Turin.
Link to section 'Accounts on Gautschi' of 'Accounts' Accounts on Gautschi
Link to section 'Obtaining an Account' of 'Accounts' Obtaining an Account
To obtain an account, you must be part of a research group which has purchased access to Gautschi. Refer to the Accounts / Access page for more details on how to request access.
Link to section 'Outside Collaborators' of 'Accounts' Outside Collaborators
A valid Purdue Career Account is required for access to any resource. If you do not currently have a valid Purdue Career Account you must have a current Purdue faculty or staff member file a Request for Privileges (R4P) before you can proceed.
Logging In
To submit jobs on Gautschi, log in to the submission host gautschi.rcac.purdue.edu via SSH. This submission host is actually 8 front-end hosts: login00.gautschi through login07.gautschi. The login process randomly assigns one of these front-ends to each login to gautschi.rcac.purdue.edu.
Purdue Login
Link to section 'SSH' of 'Purdue Login' SSH
- SSH to the cluster as usual.
- When asked for a password, type your password followed by "
,push". - Your Purdue Duo client will receive a notification to approve the login.
Link to section 'Thinlinc' of 'Purdue Login' Thinlinc
- When asked for a password, type your password followed by "
,push". - Your Purdue Duo client will receive a notification to approve the login.
- The native Thinlinc client will prompt for Duo approval twice due to the way Thinlinc works.
- The native Thinlinc client also supports key-based authentication.
Passwords
Gautschi supports either Purdue two-factor authentication (Purdue Login) or SSH keys.
SSH Client Software
Secure Shell or SSH is a way of establishing a secure connection between two computers. It uses public-key cryptography to authenticate the user with the remote computer and to establish a secure connection. Its usual function involves logging in to a remote machine and executing commands. There are many SSH clients available for all operating systems:
Linux / Solaris / AIX / HP-UX / Unix:
- The
sshcommand is pre-installed. Log in usingssh myusername@gautschi.rcac.purdue.edufrom a terminal.
Microsoft Windows:
- MobaXterm is a small, easy to use, full-featured SSH client. It includes X11 support for remote displays, SFTP capabilities, and limited SSH authentication forwarding for keys.
Mac OS X:
- The
sshcommand is pre-installed. You may start a local terminal window from "Applications->Utilities". Log in by typing the commandssh myusername@gautschi.rcac.purdue.edu.
When prompted for password, enter your Purdue career account password followed by ",push ". Your Purdue Duo client will then receive a notification to approve the login.
SSH Keys
Link to section 'General overview' of 'SSH Keys' General overview
To connect to Gautschi using SSH keys, you must follow three high-level steps:
- Generate a key pair consisting of a private and a public key on your local machine.
- Copy the public key to the cluster and append it to
$HOME/.ssh/authorized_keysfile in your account. - Test if you can ssh from your local computer to the cluster without using your Purdue password.
Detailed steps for different operating systems and specific SSH client softwares are give below.
Link to section 'Mac and Linux:' of 'SSH Keys' Mac and Linux:
-
Run
ssh-keygenin a terminal on your local machine. You may supply a filename and a passphrase for protecting your private key, but it is not mandatory. To accept the default settings, press Enter without specifying a filename.
Note: If you do not protect your private key with a passphrase, anyone with access to your computer could SSH to your account on Gautschi. -
By default, the key files will be stored in
~/.ssh/id_rsaand~/.ssh/id_rsa.pubon your local machine. -
Copy the contents of the public key into
$HOME/.ssh/authorized_keyson the cluster with the following command. When asked for a password, type your password followed by ",push". Your Purdue Duo client will receive a notification to approve the login.ssh-copy-id -i ~/.ssh/id_rsa.pub myusername@gautschi.rcac.purdue.eduNote: use your actual Purdue account user name.
If your system does not have the
ssh-copy-idcommand, use this instead:cat ~/.ssh/id_rsa.pub | ssh myusername@gautschi.rcac.purdue.edu "mkdir -p ~/.ssh && chmod 700 ~/.ssh && cat >> ~/.ssh/authorized_keys" -
Test the new key by SSH-ing to the server. The login should now complete without asking for a password.
-
If the private key has a non-default name or location, you need to specify the key by
ssh -i my_private_key_name myusername@gautschi.rcac.purdue.edu
Link to section 'Windows:' of 'SSH Keys' Windows:
| Programs | Instructions |
|---|---|
| MobaXterm | Open a local terminal and follow Linux steps |
| Git Bash | Follow Linux steps |
| Windows 10 PowerShell | Follow Linux steps |
| Windows 10 Subsystem for Linux | Follow Linux steps |
| PuTTY | Follow steps below |
PuTTY:
-
Launch PuTTYgen, keep the default key type (RSA) and length (2048-bits) and click Generate button.
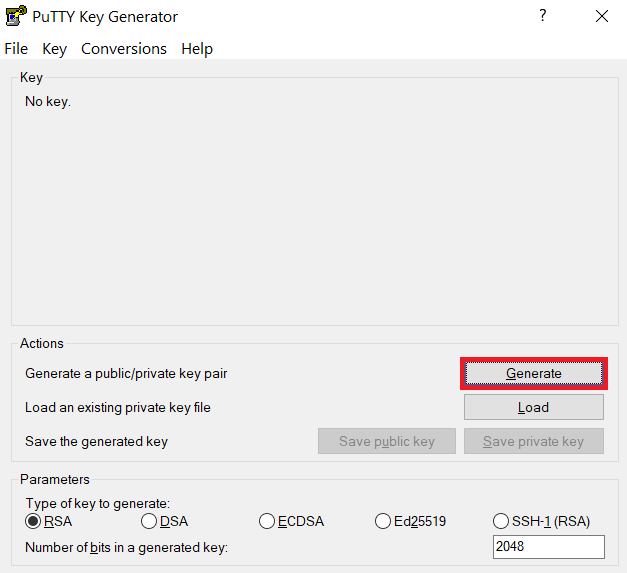
The "Generate" button can be found under the "Actions" section of the PuTTY Key Generator interface. -
Once the key pair is generated:
Use the Save public key button to save the public key, e.g.
Documents\SSH_Keys\mylaptop_public_key.pubUse the Save private key button to save the private key, e.g.
Documents\SSH_Keys\mylaptop_private_key.ppk. When saving the private key, you can also choose a reminder comment, as well as an optional passphrase to protect your key, as shown in the image below. Note: If you do not protect your private key with a passphrase, anyone with access to your computer could SSH to your account on Gautschi.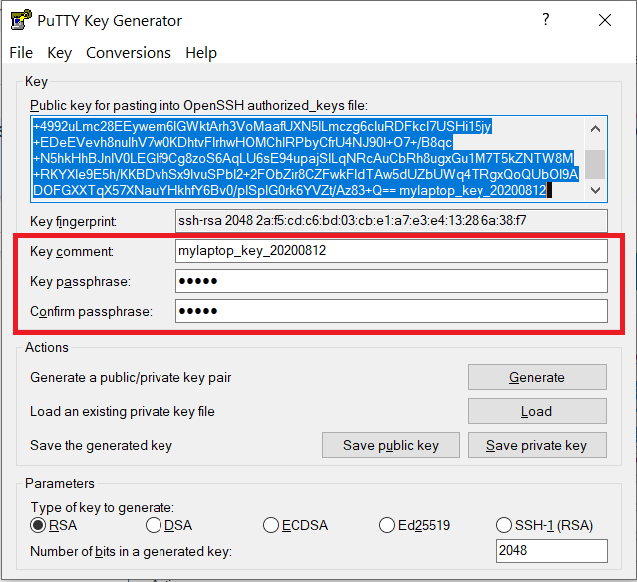
The PuTTY Key Generator form has inputs for the Key passphrase and optional reminder comment. From the menu of PuTTYgen, use the "Conversion -> Export OpenSSH key" tool to convert the private key into openssh format, e.g.
Documents\SSH_Keys\mylaptop_private_key.opensshto be used later for Thinlinc. -
Configure PuTTY to use key-based authentication:
Launch PuTTY and navigate to "Connection -> SSH ->Auth" on the left panel, click Browse button under the "Authentication parameters" section and choose your private key, e.g. mylaptop_private_key.ppk
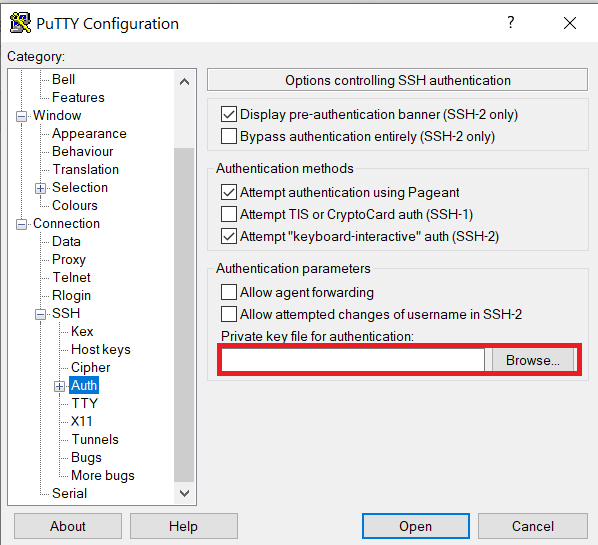
After clicking Connection -> SSH ->Auth panel, the "Browse" option can be found at the bottom of the resulting panel. Navigate back to "Session" on the left panel. Highlight "Default Settings" and click the "Save" button to ensure the change in place.
-
Connect to the cluster. When asked for a password, type your password followed by "
,push". Your Purdue Duo client will receive a notification to approve the login. Copy the contents of public key from PuTTYgen as shown below and paste it into$HOME/.ssh/authorized_keys. Please double-check that your text editor did not wrap or fold the pasted value (it should be one very long line).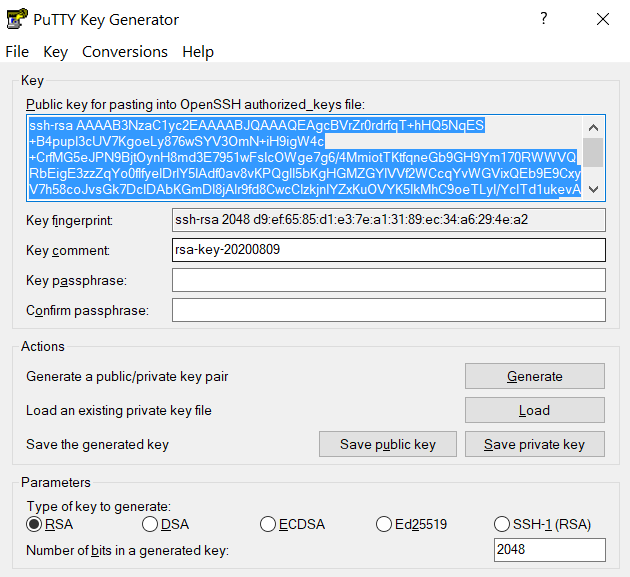
The "Public key" will look like a long string of random letters and numbers in a text box at the top of the window. - Test by connecting to the cluster. If successful, you will not be prompted for a password or receive a Duo notification. If you protected your private key with a passphrase in step 2, you will instead be prompted to enter your chosen passphrase when connecting.
SSH X11 Forwarding
SSH supports tunneling of X11 (X-Windows). If you have an X11 server running on your local machine, you may use X11 applications on remote systems and have their graphical displays appear on your local machine. These X11 connections are tunneled and encrypted automatically by your SSH client.
Link to section 'Installing an X11 Server' of 'SSH X11 Forwarding' Installing an X11 Server
To use X11, you will need to have a local X11 server running on your personal machine. Both free and commercial X11 servers are available for various operating systems.
Linux / Solaris / AIX / HP-UX / Unix:
- An X11 server is at the core of all graphical sessions. If you are logged in to a graphical environment on these operating systems, you are already running an X11 server.
- ThinLinc is an alternative to running an X11 server directly on your Linux computer. ThinLinc is a service that allows you to connect to a persistent remote graphical desktop session.
Microsoft Windows:
- ThinLinc is an alternative to running an X11 server directly on your Windows computer. ThinLinc is a service that allows you to connect to a persistent remote graphical desktop session.
- MobaXterm is a small, easy to use, full-featured SSH client. It includes X11 support for remote displays, SFTP capabilities, and limited SSH authentication forwarding for keys.
Mac OS X:
- X11 is available as an optional install on the Mac OS X install disks prior to 10.7/Lion. Run the installer, select the X11 option, and follow the instructions. For 10.7+ please download XQuartz.
- ThinLinc is an alternative to running an X11 server directly on your Mac computer. ThinLinc is a service that allows you to connect to a persistent remote graphical desktop session.
Link to section 'Enabling X11 Forwarding in your SSH Client' of 'SSH X11 Forwarding' Enabling X11 Forwarding in your SSH Client
Once you are running an X11 server, you will need to enable X11 forwarding/tunneling in your SSH client:
ssh: X11 tunneling should be enabled by default. To be certain it is enabled, you may usessh -Y.- MobaXterm: Select "New session" and "SSH." Under "Advanced SSH Settings" check the box for X11 Forwarding.
SSH will set the remote environment variable $DISPLAY to "localhost:XX.YY" when this is working correctly. If you had previously set your $DISPLAY environment variable to your local IP or hostname, you must remove any set/export/setenv of this variable from your login scripts. The environment variable $DISPLAY must be left as SSH sets it, which is to a random local port address. Setting $DISPLAY to an IP or hostname will not work.
ThinLinc
RCAC provides Cendio's ThinLinc as an alternative to running an X11 server directly on your computer. It allows you to run graphical applications or graphical interactive jobs directly on Gautschi through a persistent remote graphical desktop session.
ThinLinc is a service that allows you to connect to a persistent remote graphical desktop session. This service works very well over a high latency, low bandwidth, or off-campus connection compared to running an X11 server locally. It is also very helpful for Windows users who do not have an easy to use local X11 server, as little to no set up is required on your computer.
There are two ways in which to use ThinLinc: preferably through the native client or through a web browser.
Link to section 'Installing the ThinLinc native client' of 'ThinLinc' Installing the ThinLinc native client
The native ThinLinc client will offer the best experience especially over off-campus connections and is the recommended method for using ThinLinc. It is compatible with Windows, Mac OS X, and Linux.
- Download the ThinLinc client from the ThinLinc website.
- Start the ThinLinc client on your computer.
- In the client's login window, use desktop.gautschi.rcac.purdue.edu as the Server. Use your Purdue Career Account username and password, but append "
,push" to your password. - Click the Connect button.
- Your Purdue Login Duo will receive a notification to approve your login.
- Continue to following section on connecting to Gautschi from ThinLinc.
Link to section 'Using ThinLinc through your web browser' of 'ThinLinc' Using ThinLinc through your web browser
The ThinLinc service can be accessed from your web browser as a convenience to installing the native client. This option works with no set up and is a good option for those on computers where you do not have privileges to install software. All that is required is an up-to-date web browser. Older versions of Internet Explorer may not work.
- Open a web browser and navigate to desktop.gautschi.rcac.purdue.edu.
- Log in with your Purdue Career Account username and password, but append "
,push" to your password. - You may safely proceed past any warning messages from your browser.
- Your Purdue Login Duo will receive a notification to approve your login.
- Continue to the following section on connecting to Gautschi from ThinLinc.
Link to section 'Connecting to Gautschi from ThinLinc' of 'ThinLinc' Connecting to Gautschi from ThinLinc
- Once logged in, you will be presented with a remote Linux desktop running directly on a cluster front-end.
- Open the terminal application on the remote desktop.
- Once logged in to the Gautschi head node, you may use graphical editors, debuggers, software like Matlab, or run graphical interactive jobs. For example, to test the X forwarding connection issue the following command to launch the graphical editor gedit:
$ gedit - This session will remain persistent even if you disconnect from the session. Any interactive jobs or applications you left running will continue running even if you are not connected to the session.
Link to section 'Tips for using ThinLinc native client' of 'ThinLinc' Tips for using ThinLinc native client
- To exit a full screen ThinLinc session press the F8 key on your keyboard (fn + F8 key for Mac users) and click to disconnect or exit full screen.
- Full screen mode can be disabled when connecting to a session by clicking the Options button and disabling full screen mode from the Screen tab.
Link to section 'Configure ThinLinc to use SSH Keys' of 'ThinLinc' Configure ThinLinc to use SSH Keys
- The web client does NOT support public-key authentication.
-
ThinLinc native client supports the use of an SSH key pair. For help generating and uploading keys to the cluster, see SSH Keys section in our user guide for details.
To set up SSH key authentication on the ThinLinc client:
-
Open the Options panel, and select Public key as your authentication method on the Security tab.
The "Options..." button in the ThinLinc Client can be found towards the bottom left, above the "Connect" button. -
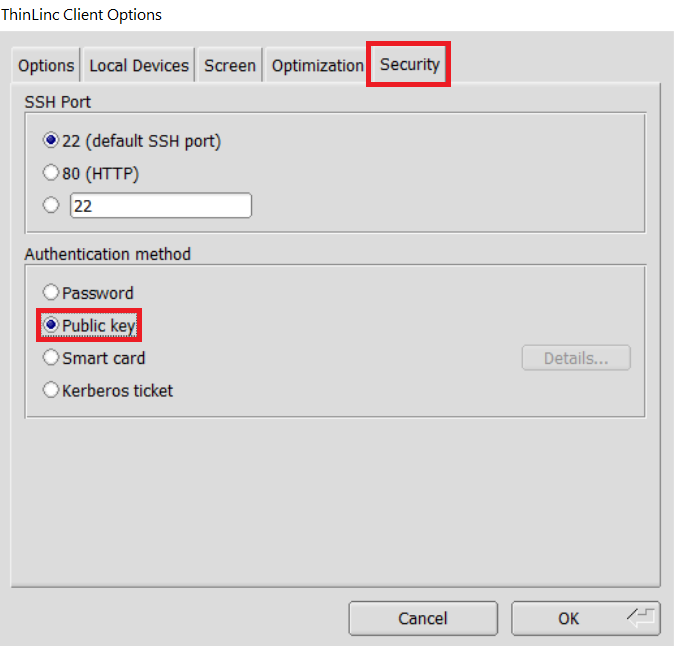
In the options dialog, switch to the "Security" tab and select the "Public key" radio button:
The "Security" tab found in the options dialog, will be the last of available tabs. The "Public key" option can be found in the "Authentication method" options group. - Click OK to return to the ThinLinc Client login window. You should now see a Key field in place of the Password field.
-
In the Key field, type the path to your locally stored private key or click the ... button to locate and select the key on your local system. Note: If PuTTY is used to generate the SSH Key pairs, please choose the private key in the openssh format.
The ThinLinc Client login window will now display key field instead of a password field.
-
Software
Link to section 'Environment module' of 'Software' Environment module
Link to section 'Software catalog' of 'Software' Software catalog
Our clusters provide a number of software packages to users of the system via the module command.
Link to section 'Environment Management with the Module Command' of 'Environment Management with the Module Command' Environment Management with the Module Command
The module command is the preferred method to manage your processing environment. With this command, you may load applications and compilers along with their libraries and paths. Modules are packages that you load and unload as needed.
Please use the module command and do not manually configure your environment, as staff may make changes to the specifics of various packages. If you use the module command to manage your environment, these changes will not be noticeable.
Link to section 'Hierarchy' of 'Environment Management with the Module Command' Hierarchy
Many modules have dependencies on other modules. For example, a particular openmpi module requires a specific version of the Intel compiler to be loaded. Often, these dependencies are not clear to users of the module, and there are many modules which may conflict. Arranging modules in a hierarchical fashion makes this dependency clear. This arrangement also helps make the software stack easy to understand - your view of the modules will not be cluttered with a bunch of conflicting packages.
Your default module view on Gautschi will include a set of compilers and a set of basic software that has no dependencies (such as Matlab and Fluent). To make software available that depends on a compiler, you must first load the compiler, and then software which depends on it becomes available to you. In this way, all software you see when doing module avail is completely compatible with each other.
Link to section 'Using the Hierarchy' of 'Environment Management with the Module Command' Using the Hierarchy
Your default module view on Gautschi will include a set of compilers and a set of basic software that has no dependencies (such as Matlab and Fluent).
To see what modules are available on this system by default:
$ module avail
To see which versions of a specific compiler are available on this system:
$ module avail gcc
$ module avail intel
To continue further into the hierarchy of modules, you will need to choose a compiler. As an example, if you are planning on using the Intel compiler you will first want to load the Intel compiler:
$ module load intel
With intel loaded, you can repeat the avail command, and at the bottom of the output you will see the section of additional software that the intel module provides:
$ module avail
Several of these new packages also provide additional software packages, such as MPI libraries. You can repeat the last two steps with one of the MPI packages such as openmpi and you will have a few more software packages available to you.
If you are looking for a specific software package and do not see it in your default view, the module command provides a search function for searching the entire hierarchy tree of modules without the need for you to manually load and avail on every module.
To search for a software package:
$ module spider openmpi
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
openmpi:
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Versions:
openmpi/4.1.6
openmpi/5.0.5
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
For detailed information about a specific "openmpi" package (including how to load the modules) use the module's full name.
Note that names that have a trailing (E) are extensions provided by other modules.
For example:
$ module spider openmpi/5.0.5
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
This will search for the openmpi software package. If you do not specify a specific version of the package, you will be given a list of versions available on the system. Select the version you wish to use and spider that to see how to access the module:
$ module spider openmpi/5.0.5
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
openmpi: openmpi/5.0.5
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
You will need to load all module(s) on any one of the lines below before the "openmpi/5.0.5" module is available to load.
gcc/11.4.1
gcc/14.1.0
Help:
An open source Message Passing Interface implementation. The Open MPI
Project is an open source Message Passing Interface implementation that
is developed and maintained by a consortium of academic, research, and
industry partners. Open MPI is therefore able to combine the expertise,
technologies, and resources from all across the High Performance
Computing community in order to build the best MPI library available.
Open MPI offers advantages for system and software vendors, application
developers and computer science researchers.
The output of this command will instruct you that you can load this module directly, or in the case of the above example, that you will need to first load a module or two. With the information provided with this command, you can now construct a load command to load a version of OpenMPI into your environment:
$ module load gcc/14.1.0 openmpi/5.0.5
Some user communities may maintain copies of their domain software for others to use. For example, the Purdue Bioinformatics Core provides a wide set of bioinformatics software for use by any user of RCAC clusters via the bioinfo module. The spider command will also search this repository of modules. If it finds a software package available in the bioinfo module repository, the spider command will instruct you to load the bioinfo module first.
Link to section 'Load / Unload a Module' of 'Environment Management with the Module Command' Load / Unload a Module
All modules consist of both a name and a version number. When loading a module, you may use only the name to load the default version, or you may specify which version you wish to load.
For each cluster, RCAC makes a recommendation regarding the set of compiler, math library, and MPI library for parallel code. To load the recommended set:
$ module load rcac
To verify what you loaded:
$ module list
To load the default version of a specific compiler, choose one of the following commands:
$ module load gcc
$ module load intel
To load a specific version of a compiler, include the version number:
$ module load gcc/11.2.0
When running a job, you must use the job submission file to load on the compute node(s) any relevant modules. Loading modules on the front end before submitting your job makes the software available to your session on the front-end, but not to your job submission script environment. You must load the necessary modules in your job submission script.
To unload a compiler or software package you loaded previously:
$ module unload gcc
$ module unload intel
$ module unload matlab
To unload all currently loaded modules and reset your environment:
$ module purge
Link to section 'Show Module Details' of 'Environment Management with the Module Command' Show Module Details
To learn more about what a module does to your environment, you may use the module show command.
$ module show matlab
Here is an example showing what loading the default Matlab does to the processing environment:
-------------------------------------------------------------------------------------------------------------------------------------------
/opt/spack/modulefiles/Core/matlab/R2022a.lua:
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
whatis("Name : matlab")
whatis("Version : R2022a")
...
setenv("MATLAB_HOME","/apps/spack/gautschi/apps/matlab/R2022a-gcc-8.5.0-u54n6sa")
setenv("RCAC_MATLAB_ROOT","/apps/spack/gautschi/apps/matlab/R2022a-gcc-8.5.0-u54n6sa")
setenv("RCAC_MATLAB_VERSION","R2022a")
setenv("MATLAB","/apps/spack/gautschi/apps/matlab/R2022a-gcc-8.5.0-u54n6sa")
setenv("MLROOT","/apps/spack/gautschi/apps/matlab/R2022a-gcc-8.5.0-u54n6sa")
setenv("ARCH","glnxa64")
append_path("PATH","/apps/spack/gautschi/apps/matlab/R2022a-gcc-8.5.0-u54n6sa/bin/glnxa64:/apps/spack/gautschi/apps/matlab/R2019a-gcc-4.8.5-jg35hvf/bin")
append_path("CMAKE_PREFIX_PATH","/apps/spack/gautschi/apps/matlab/R2022a-gcc-8.5.0-u54n6sa/")
append_path("LD_LIBRARY_PATH","/apps/spack/gautschi/apps/matlab/R2022a-gcc-8.5.0-u54n6sa/runtime/glnxa64:/apps/spack/gautschi/apps/matlab/R2022a-gcc-8.5.0-u54n6sa/bin/glnxa64")
Link to section 'GPU modules' of 'Environment Management with the Module Command' GPU modules
We use different module branch for GPU applications on Gautschi. To use the GPU modules, you can module load the gpu module branch first:
$ module load modtree/gpu
After loading the modtree/gpu, you are able to use module spider to search modules or module load to add the desired module to your environment.
For more information about Lmod:
Running Jobs
There is one method for submitting jobs to Gautschi. You may use SLURM to submit jobs to a partition on Gautschi. SLURM performs job scheduling. Jobs may be any type of program. You may use either the batch or interactive mode to run your jobs. Use the batch mode for finished programs; use the interactive mode only for debugging.
In this section, you'll find a few pages describing the basics of creating and submitting SLURM jobs. As well, a number of example SLURM jobs that you may be able to adapt to your own needs.
Basics of SLURM Jobs
The Simple Linux Utility for Resource Management (SLURM) is a system providing job scheduling and job management on compute clusters. With SLURM, a user requests resources and submits a job to a queue. The system will then take jobs from queues, allocate the necessary nodes, and execute them.
Do NOT run large, long, multi-threaded, parallel, or CPU-intensive jobs on a front-end login host. All users share the front-end hosts, and running anything but the smallest test job will negatively impact everyone's ability to use Gautschi. Always use SLURM to submit your work as a job.
Link to section 'Submitting a Job' of 'Basics of SLURM Jobs' Submitting a Job
The main steps to submitting a job are:
Follow the links below for information on these steps, and other basic information about jobs. A number of example SLURM jobs are also available.
Queues
On Gautschi, the required options for job submisison deviates from some of the other community clusters you might have experience using. In general every job submission will have four parts:
- The number and type of resources you want
- The account the resources should come out of
- The quality of service (QOS) this job expects from the resources
- The partition where the resources are located
If you have used other clusters, you will be familiar with first item. If you have not, you can read about how to format the request on our job submission page. The rest of this page will focus on the last three items as they are specific to Gautschi.
Link to section 'Accounts' of 'Queues' Accounts
On the Gautschi community cluster, users will have access to one or more accounts, also known as queues. These accounts are dedicated to and named after each partner who has purchased access to the cluster, and they provide partners and their researchers with priority access to their portion of the cluster. These accounts can be thought of as bank accounts that contain the resources a group has purchased access to which may include some number of cores, GPU hours or both. To see the list of accounts that you have access to on Gautschi as well as the resources they contain, you can use the command "slist".
On previous clusters, you may have had access to several other special accounts like "standby", "highmem", "gpu", etc. On Gautschi, these modes of access still exist, but they are no longer accounts. As well on Gautschi, you must explicitly define the account that you want to submit to using the "-A/--account=" option.
Link to section 'Quality of Service (QOS)' of 'Queues' Quality of Service (QOS)
On Gautschi, we use a Slurm concept called a Quality of Service or a QOS. A QOS can be thought of as a tag for a job that tells the scheduler how that job should be treated with respect to limits, priority, etc. The cluster administrators define the available QOSes as well as the policies for how each QOS should be treated on the cluster. A toy example of such a policy may be "no single user can have more than 200 jobs" that has been tagged with a QOS named "highpriority". There are two classes of QOSes and a job can have both:
- Partition QOSes: A partition QOS is a tag that is automatically added to your job when you submit to a partition that defines a partition QOS.
- Job QOSes: A Job QOS is a tag that you explicitly give to a job using the option "-q/--qos=". By explicitly tagging your jobs this way, you can choose the policy that each one of your jobs should abide by. We will describe the policies for the available job QOSes in the partition section below.
As an extended metaphor, if we think of a job as a package that we need to have shipped to some destination, then the partition can be thought of as the carrier we decide to ship our package with. That carrier is going to have some company policies that dictate how you need to label/pack that package, and that company policy is like the partition QOS. It is the policy that is enforced for simply deciding to submit to a particular partition. The Job QOS can then be thought of as the various different types of shipping options that carrier might offer. You might pay extra to have that package shipped overnight or have tracking information for it or you may choose to pay less and have your package arrive as available. Once we decide to go with a particular carrier, we are subject to their company policy, but we may negotiate how they handle that package by choosing one of their available shipping options. In the same way, when you choose to submit to a partition, you are subject to the limits enforced by the partition QOS and you may be able to ask for your job to be handled a particular way by specifying the job QOS, but that option has to be offered by the partition.
In order for a job to use a Job QOS, the user submitting the job must have access to the QOS, the account the job is being submitted to must accept the QOS, and the partition the job is being submitted to must accept the QOS. The following job QOSes every user and every account of Gautschi has access to:
- "normal": The "normal" QOS is the default job QOS on the cluster meaning if you do not explicitly list an alternative job QOS, your job will be tagged with this QOS. The policy for this QOS provides a high priority and does not add any additional limits.
- "standby": The "standby" QOS must be explicitly used if desired by using the option "-q standby" or "--qos=standby". The policy for this QOS gives access to idle resources on the cluster. Jobs tagged with this QOS are "low priority" jobs and are only allowed to run for up to four hours at a time, however the resources used by these jobs do not count against the resources in your Account. For users of our previous clusters, usage of this QOS replaces the previous -A standby style of submission.
Some of these QOSes may not be available in every partition. Each of the partitions in the following section will enumerate which of these QOSes are allowed in the partition.
Link to section 'Partitions' of 'Queues' Partitions
On Gautschi, the various types of nodes on the cluster are organized into distinct partitions. This allows CPU and GPU nodes to be charged separately and differently. This also means that Instead of only needing to specify the account name in the job script, the desired partition must also be specified. Each of these partitions is subject to different limitations and has a specific use case that will be described below.
Link to section 'CPU Partition' of 'Queues' CPU Partition
This partition contains the resources a group purchases access to when they purchase CPU resources on Gautschi and is made up of 336 Gautschi-A nodes. Each of these nodes contains two Zen 4 AMD EPYC 9654 96-core processors for a total of 192 cores and 384 GB of memory for a total of more than 64,000 cores in the partition. Memory in this partition is allocated proportional to your core request such that each core is given about 2 GB of memory per core requested. Submission to this partition can be accomplished by using the option: "-p cpu" or "--partition=cpu".
The purchasing model for this partition allows groups to purchase high priority access to some number of cores. When an account uses resources in this account by submitting a job tagged with the "normal" QOS, the cores used by that job are withdrawn from the account and deposited back into the account when the job terminates.
When using the CPU partition, jobs are tagged by the "normal" QOS by default, but they can be tagged with the "standby" QOS if explicitly submitted using the "-q standby" or "--qos=standby" option.
- Jobs tagged with the "normal" QOS are subject to the following policies:
- Jobs have a high priority and should not need to wait very long before starting.
- Any cores requested by these jobs are withdrawn from the account until the job terminates.
- These jobs can run for up to two weeks at a time.
- Jobs tagged with the "standby" QOS are subject to the following policies:
- Jobs have a low priority and there is no expectation of job start time. If the partition is very busy with jobs using the "normal" QOS or if you are requesting a very large job, then jobs using the "standby" QOS may take hours or days to start.
- These jobs can use idle resources on the cluster and as such cores requested by these jobs are not withdrawn from the account to which they were submitted.
- These jobs can run for up to four hours at a time.
Groups who purchased GPU resources on Gautschi will have the ability to run jobs in this partition tagged with the "standby" QOS described above, but will not be able to submit jobs tagged with the "normal" qos unless they have purchased CPU resources on the cluster.
Available QOSes: "normal", "standby"
Link to section 'AI Partition' of 'Queues' AI Partition
This partition contains the resources a group purchases access to when they purcahse GPU resources on Gautschi and is made up of 20 Gautschi-H nodes. Each of these nodes contains two Intel Xeon Platinum 8480+ 56-core processors and 8-way NVLinked H100s for a total of 160 H100s in the partition. CPU memory in this partition is allocated proportional to your core request such that each core is given about 9 GB of memory per core requested. Submission to this partition can be accomplished by using the option: "-p ai" or "--partition=ai".
The purchasing model for this cluster allows groups to purchase access to GPUs. When a group purchases access to a GPU for a year, they are given 365*24 hours of GPU time on the partition. The advantage to tracking resources this way is that if a group needs access to more than one GPU for a project, they can use multiple GPUs at once since instead of having to purchase additional GPUs, they just consume those GPU hours faster. When an account uses resources in this partition, the balance of GPU hours in their account is charged at a rate of at least one GPU hour for each GPU their job uses per hour (this is tracked by the minute), however the usage of some QOSes described below will charge usage at a slower rate.
When using the AI partition, jobs are tagged by the "normal" QOS by default, but they can be tagged with the "preemptible" QOS if explicitly submitted using the "-q preemptible" or "--qos=preemptible" option.
- Jobs tagged with the "normal" QOS are subject to the following policies:
- Jobs have a high priority and should not need to wait very long before starting.
- The account the job is submitted to will be charged 1 GPU hour for each GPU this job uses per hour (this is tracked by the minute).
- These jobs can run for up to two weeks at a time.
- Jobs tagged with the "preemptible" QOS subject to the following policies:
- Jobs have a low priority and there is no expectation of job start time. If the partition is very busy with jobs using the "normal" QOS or if you are requesting a very large job, then jobs using the "standby" QOS may take hours or days to start.
- The account the job is submitted to will be charged 0.25 GPU hour for each GPU this job uses per hour (this is tracked by the minute) which effectively allows jobs to use 4x as many resources as a job tagged with the "normal" qos for the same cost.
- These jobs can run for up to two weeks at a time.
- If there are not enough idle resources in this partition for a job tagged with the "normal" QOS to start, then this job may be cancelled to make room for that job. This means it is imperative to use checkpointing if using this QOS.
Available QOSes: "normal", "preemptible"
Link to section 'Highmem Partition' of 'Queues' Highmem Partition
This partition is made up of 6 Gautschi-B nodes which have four times as much memory as a standard Gautschi-A node, and access to this partition is given to all accounts on the cluster to enable work that has higher memory requirements. Each of these nodes contains two Zen 4 AMD EPYC 9654 96-core processors for a total of 192 cores and 1.5 TB of memory. Memory in this partition is allocated proportional to your core request such that each core is given about 8 GB of memory per core requested. Submission to this partition can be accomplished by using the option: "-p highmem" or "--partition=highmem".
When using the Highmem partition, jobs are tagged by the "normal" QOS by default, and this is the only QOS that is available for this partition, so there is no need to specify a QOS when using this partition. Additionally jobs are tagged by a highmem partition QOS that enforces the following policies
- There is no expectation of job start time as these nodes are a shared resources that are given as a bonus for purchasing access to high priority access to resources on Gautschi
- You can have 2 jobs running in this partition at once
- You can have 8 jobs submitted to thie partition at once
- Your jobs must use more than 48 of the 192 cores on the node otherwise your memory footprint would fit on a standard Gautschi-A node
- These jobs can run for up to 24 hours at a time.
Available QOSes: "normal"
Link to section 'Profiling Partition' of 'Queues' Profiling Partition
This partition is made up of 2 Gautschi-A nodes that have hardware performance counters enabled. By enabling hardware performance counters, profiling applications such as AMD MicroProf can track certain performance criteria for execution on the CPU such as L3 cache events, speculative execution misses, etc. Due to the fact that this allows greater visibility into the execution of each process, the nodes in this partition can only be used if your job uses the entire node. Submission to this partition can be accomplished by using the option: "-p profiling" or "--partition=profiling".
When using the profiling partition, jobs are tagged by the "normal" QOS by default, and this is the only QOS that is available for this partition, so there is no need to specify a QOS when using this partition. Additionally jobs are tagged by a profiling partition QOS that enforces the following policies
- There is no expectation of job start time as these nodes are a shared resources that are given as a bonus for purchasing access to high priority access to resources on Gautschi
- You can have 1 job running in this partition at once
- If you have a single process running on one of the nodes, you must use that entire node when submitting to this partition
- These jobs can run for up to 24 hours at a time.
This is a resource that is reserved for groups profiling their applications and we monitor to ensure that this partition is not being used simply for more compute.
Available QOSes: "normal"
Link to section 'Smallgpu Partiton' of 'Queues' Smallgpu Partiton
This partition is made up of 6 Gautschi-G nodes. Each of these nodes contains two NVIDIA L40s and two Zen 4 AMD EPYC 9554 64-core processors for a total of 128 cores and 384GB of memory. Memory in this partition is allocated proportional to your core request such that each core is given about 3 GB of memory per core requested. You should request cores proportional to the number of GPUs you are using in this partition (i.e. if you only need one of the two GPUs, you should request half of the cores on the node) Submission to this partition can be accomplished by using the option: "-p smallgpu" or "--partition=smallgpu".
When using the smallgpu partition, jobs are tagged by the "normal" QOS by default, and this is the only QOS that is available for this partition, so there is no need to specify a QOS when using this partition. Additionally jobs are tagged by a smallgpu partition QOS that enforces the following policies
- There is no expectation of job start time as these nodes are a shared resources that are given as a bonus for purchasing access to high priority access to resources on Gautschi
- You can use up to 2 GPUs in this partition at once
- You can have 8 jobs submitted to thie partition at once
- These jobs can run for up to 24 hours at a time.
Available QOSes: "normal"
Job Submission Script
To submit work to a SLURM queue, you must first create a job submission file. This job submission file is essentially a simple shell script. It will set any required environment variables, load any necessary modules, create or modify files and directories, and run any applications that you need:
#!/bin/bash
# FILENAME: myjobsubmissionfile
# Loads Matlab and sets the application up
module load matlab
# Change to the directory from which you originally submitted this job.
cd $SLURM_SUBMIT_DIR
# Runs a Matlab script named 'myscript'
matlab -nodisplay -singleCompThread -r myscript
Once your script is prepared, you are ready to submit your job.
Link to section 'Job Script Environment Variables' of 'Job Submission Script' Job Script Environment Variables
| Name | Description |
|---|---|
| SLURM_SUBMIT_DIR | Absolute path of the current working directory when you submitted this job |
| SLURM_JOBID | Job ID number assigned to this job by the batch system |
| SLURM_JOB_NAME | Job name supplied by the user |
| SLURM_JOB_NODELIST | Names of nodes assigned to this job |
| SLURM_CLUSTER_NAME | Name of the cluster executing the job |
| SLURM_SUBMIT_HOST | Hostname of the system where you submitted this job |
| SLURM_JOB_PARTITION | Name of the original queue to which you submitted this job |
Submitting a Job
Once you have a job submission file, you may submit this script to SLURM using the sbatch command. SLURM will find, or wait for, available resources matching your request and run your job there.
On Gautschi, in order to submit jobs, you need to specify the partition, account and Quality of Service (QoS) name to which you want to submit your jobs. To familiarize yourself with the partitions and QoS available on Gautschi, visit Gautschi Queues and Partitons. To check the available partitions on Gautschi, you can use the showpartitions , and to check your available accounts you can use slist commands. Slurm uses the term "Account" with the option -A or --account= to specify different batch accounts, the option -p or --partition= to select a specific partition for job submission, and the option -q or --qos= .
showpartitions
Partition statistics for cluster gautschi at Fri Dec 6 05:26:30 PM EST 2024
Partition #Nodes #CPU_cores Cores_pending Job_Nodes MaxJobTime Cores Mem/Node
Name State Total Idle Total Idle Resorc Other Min Max Day-hr:mn /node (GB)
ai up 20 20 2240 2240 0 0 1 infin infinite 112 1031
cpu up 336 300 64512 57600 0 0 1 infin infinite 192 386
highmem up 6 6 1152 1152 0 0 1 infin infinite 192 1547
smallgpu up 6 6 768 768 0 0 1 infin infinite 128 386
profiling up 2 2 384 384 0 0 1 infin infinite 192 386
Link to section 'CPU Partition' of 'Submitting a Job' CPU Partition
The CPU partition on Gautschi has two Quality of Service (QoS) levels: normal and standby. To submit your job to one compute node on cpu partition and 'normal' QoS which has "high priority":
$ sbatch --nodes=1 --ntasks=1 --partition=cpu --account=accountname --qos=normal myjobsubmissionfile
$ sbatch -N1 -n1 -p cpu -A accountname -q normal myjobsubmissionfile
To submit your job to one compute node on cpu partition and 'standby' QoS which is has "low priority":
$ sbatch --nodes=1 --ntasks=1 --partition=cpu --account=accountname --qos=standby myjobsubmissionfile
$ sbatch -N1 -n1 -p cpu -A accountname -q standby myjobsubmissionfile
Link to section ' AI Partition' of 'Submitting a Job' AI Partition
The CPU partition on Gautschi has two Quality of Service (QoS) levels: normal and preemptible. To submit your job to one compute node requesting one GPU on ai partition and 'normal' QoS which is has "high priority":
$ sbatch --nodes=1 --gpus-per-node=1 --ntasks=14 --partition=ai --account=accountname --qos=normal myjobsubmissionfile
$ sbatch -N1 --gpus-per-node=1 -n14 -p ai -A accountname -q normal myjobsubmissionfile
To submit your job to one compute node requesting one GPU on ai partition and 'preemptible' QoS which has "high priority":
$ sbatch --nodes=1 --gpus-per-node=1 --ntasks=14 --partition=ai --account=accountname --qos=preemptible myjobsubmissionfile
$ sbatch -N1 --gpus-per-node=1 -n14 -p ai -A accountname -q preemptible myjobsubmissionfile
Link to section 'Highmem Partition' of 'Submitting a Job' Highmem Partition
To submit your job to a compute node on the highmem partition, you don’t need to specify the QoS name because only one QoS exists for this partition, and the default is normal. However, the highmem partition is suitable for jobs with memory requirements that exceed the capacity of a standard node, so the number of requested tasks should be appropriately high.
Link to section 'Profiling Partition' of 'Submitting a Job' Profiling Partition
To submit your job to a compute node on the profiling partition, you also don’t need to specify the QoS name because only one QoS exists for this partition, and the default is normal.
$ sbatch --nodes=1 --ntasks=1 --partition=profiling --account=accountname myjobsubmissionfile
$ sbatch -N1 -n1 -p profiling -A accountname myjobsubmissionfile
Link to section 'Smallgpu Partition' of 'Submitting a Job' Smallgpu Partition
To submit your job to a compute node on the smallgpu partition, you don’t need to specify the QoS name because only one QoS exists for this partition, and the default is normal. You should request cores proportional to the number of GPUs you are using in this partition (i.e. if you only need one of the two GPUs, you should request half of the cores on the node).
$ sbatch --nodes=1 --ntasks=64 --gpus-per-node=1 --partition=smallgpu --account=accountname myjobsubmissionfile
$ sbatch -N1 -n64 --gpus-per-node=1 -p smallgpu -A accountname myjobsubmissionfile
Link to section 'General Information' of 'Submitting a Job' General Information
By default, each job receives 30 minutes of wall time, or clock time. If you know that your job will not need more than a certain amount of time to run, request less than the maximum wall time, as this may allow your job to run sooner. To request the 1 hour and 30 minutes of wall time:
$ sbatch -t 01:30:00 -N=1 -n=1 -p=cpu -A=accountname -q=standby myjobsubmissionfile
The --nodes= or -N value indicates how many compute nodes you would like for your job, and --ntasks= or -n value indicates the number of tasks you want to run.
In some cases, you may want to request multiple nodes. To utilize multiple nodes, you will need to have a program or code that is specifically programmed to use multiple nodes such as with MPI. Simply requesting more nodes will not make your work go faster. Your code must support this ability.
To request 2 compute nodes:
$ sbatch -t 01:30:00 -N=2 -n=16 -p=cpu -A=accountname -q=standby myjobsubmissionfile
By default, jobs on Gautschi will share nodes with other jobs.
If more convenient, you may also specify any command line options to sbatch from within your job submission file, using a special form of comment:
#!/bin/sh -l
# FILENAME: myjobsubmissionfile
#SBATCH --account=accountname
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --partition=cpu
#SBATCH --qos=normal
#SBATCH --time=1:30:00
#SBATCH --job-name myjobname
# Print the hostname of the compute node on which this job is running.
/bin/hostname
If an option is present in both your job submission file and on the command line, the option on the command line will take precedence.
After you submit your job with SBATCH, it may wait in queue for minutes, hours, or even weeks. How long it takes for a job to start depends on the specific queue, the resources and time requested, and other jobs already waiting in that queue requested as well. It is impossible to say for sure when any given job will start. For best results, request no more resources than your job requires.
Once your job is submitted, you can monitor the job status, wait for the job to complete, and check the job output.
Checking Job Status
Once a job is submitted there are several commands you can use to monitor the progress of the job.
To see your jobs, use the squeue -u command and specify your username:
(Remember, in our SLURM environment a queue is referred to as an 'Account')
squeue -u myusername
JOBID USER ACCOUNT PART QOS NAME NODES TRES_PER_NODE CPUS TIME_LIMIT ST TIME
182792 myusername account_name cpu standby job1 1 N/A 192 4:00:00 R 35:54
185841 myusername account_name cpu normal job2 1 N/A 192 4:00:00 R 36:51
185844 myusername account_name smal normal job3 1 gres/gpu:1 64 8:00:00 R 22:11
To retrieve useful information about your queued or running job, use the scontrol show job command with your job's ID number. The output should look similar to the following:
scontrol show job 3519
JobId=3519 JobName=job.sub
UserId=myusername GroupId=mygroup MCS_label=N/A
Priority=1000 Nice=0 Account=myaccount QOS=normal
JobState=RUNNING Reason=None Dependency=(null)
Requeue=1 Restarts=0 BatchFlag=0 Reboot=0 ExitCode=0:0
RunTime=00:00:23 TimeLimit=04:00:00 TimeMin=N/A
SubmitTime=2025-02-24T14:31:19 EligibleTime=2025-02-24T14:31:19
AccrueTime=2025-02-24T14:31:19
StartTime=2025-02-24T14:32:36 EndTime=2025-02-24T18:32:36 Deadline=N/A
PreemptEligibleTime=2025-02-24T14:32:36 PreemptTime=None
SuspendTime=None SecsPreSuspend=0 LastSchedEval=2025-02-24T14:32:36 Scheduler=Backfill
Partition=cpu AllocNode:Sid=login02:4061246
ReqNodeList=(null) ExcNodeList=(null)
NodeList=a166
BatchHost=a166
NumNodes=1 NumCPUs=4 NumTasks=4 CPUs/Task=1 ReqB:S:C:T=0:0:*:*
ReqTRES=cpu=4,mem=7668M,node=1,gres/hp_cpu=4
AllocTRES=cpu=4,mem=7668M,node=1,gres/hp_cpu=4
Socks/Node=* NtasksPerN:B:S:C=0:0:*:* CoreSpec=*
MinCPUsNode=1 MinMemoryCPU=1917M MinTmpDiskNode=0
Features=(null) DelayBoot=00:00:00
OverSubscribe=OK Contiguous=0 Licenses=(null) Network=(null)
Command=/bin/sh
WorkDir=/home/myusername
TresPerJob=gres/hp_cpu=4
There are several useful bits of information in this output.
JobStatelets you know if the job is Pending, Running, Completed, or Held.RunTime and TimeLimitwill show how long the job has run and its maximum time.SubmitTimeis when the job was submitted to the cluster.NumNodes,NumCPUs,NumTasksandCPUs/Taskare the number of Nodes, CPUs, Tasks, and CPUs per Task are shown.WorkDiris the job's working directory.
Checking Job Output
Once a job is submitted, and has started, it will write its standard output and standard error to files that you can read.
SLURM catches output written to standard output and standard error - what would be printed to your screen if you ran your program interactively. Unless you specfied otherwise, SLURM will put the output in the directory where you submitted the job in a file named slurm- followed by the job id, with the extension out. For example slurm-3509.out. Note that both stdout and stderr will be written into the same file, unless you specify otherwise.
If your program writes its own output files, those files will be created as defined by the program. This may be in the directory where the program was run, or may be defined in a configuration or input file. You will need to check the documentation for your program for more details.
Link to section 'Redirecting Job Output' of 'Checking Job Output' Redirecting Job Output
It is possible to redirect job output to somewhere other than the default location with the --error and --output directives:
#!/bin/bash
#SBATCH --output=/home/myusername/joboutput/myjob.out
#SBATCH --error=/home/myusername/joboutput/myjob.out
# This job prints "Hello World" to output and exits
echo "Hello World"
Holding a Job
Sometimes you may want to submit a job but not have it run just yet. You may be wanting to allow lab mates to cut in front of you in the queue - so hold the job until their jobs have started, and then release yours.
To place a hold on a job before it starts running, use the scontrol hold job command:
$ scontrol hold job myjobid
Once a job has started running it can not be placed on hold.
To release a hold on a job, use the scontrol release job command:
$ scontrol release job myjobid
You find the job ID using the squeue command as explained in the SLURM Job Status section.
Job Dependencies
Dependencies are an automated way of holding and releasing jobs. Jobs with a dependency are held until the condition is satisfied. Once the condition is satisfied jobs only then become eligible to run and must still queue as normal.
Job dependencies may be configured to ensure jobs start in a specified order. Jobs can be configured to run after other job state changes, such as when the job starts or the job ends.
These examples illustrate setting dependencies in several ways. Typically dependencies are set by capturing and using the job ID from the last job submitted.
To run a job after job myjobid has started:
sbatch --dependency=after:myjobid myjobsubmissionfile
To run a job after job myjobid ends without error:
sbatch --dependency=afterok:myjobid myjobsubmissionfile
To run a job after job myjobid ends with errors:
sbatch --dependency=afternotok:myjobid myjobsubmissionfile
To run a job after job myjobid ends with or without errors:
sbatch --dependency=afterany:myjobid myjobsubmissionfile
To set more complex dependencies on multiple jobs and conditions:
sbatch --dependency=after:myjobid1:myjobid2:myjobid3,afterok:myjobid4 myjobsubmissionfile
Canceling a Job
To stop a job before it finishes or remove it from a queue, use the scancel command:
scancel myjobid
You find the job ID using the squeue command as explained in the SLURM Job Status section.
Example Jobs
A number of example jobs are available for you to look over and adapt to your own needs. The first few are generic examples, and latter ones go into specifics for particular software packages.
Generic SLURM Jobs
The following examples demonstrate the basics of SLURM jobs, and are designed to cover common job request scenarios. These example jobs will need to be modified to run your application or code.
Simple Job
Every SLURM job consists of a job submission file. A job submission file contains a list of commands that run your program and a set of resource (nodes, walltime, queue) requests. The resource requests can appear in the job submission file or can be specified at submit-time as shown below.
This simple example submits the job submission file hello.sub to the cpu queue on Gautschi and requests a single node:
#!/bin/bash
# FILENAME: hello.sub
# Show this ran on a compute node by running the hostname command.
hostname
echo "Hello World"
sbatch -A myallocation -p queue-name --nodes=1 --ntasks=1 --cpus-per-task=1 --time=00:01:00 hello.sub
Submitted batch job 3521
For a real job you would replace echo "Hello World" with a command, or sequence of commands, that run your program.
After your job finishes running, the ls command will show a new file in your directory, the .out file:
ls -l
hello.sub
slurm-3521.out
The file slurm-3521.out contains the output and errors your program would have written to the screen if you had typed its commands at a command prompt:
cat slurm-3521.out
gautschi-a001.rcac.purdue.edu
Hello World
You should see the hostname of the compute node your job was executed on. Following should be the "Hello World" statement.
Multiple Node
In some cases, you may want to request multiple nodes. To utilize multiple nodes, you will need to have a program or code that is specifically programmed to use multiple nodes such as with MPI. Simply requesting more nodes will not make your work go faster. Your code must support this ability.
This example shows a request for multiple compute nodes. The job submission file contains a single command to show the names of the compute nodes allocated:
# FILENAME: myjobsubmissionfile.sub
#!/bin/bash
echo "$SLURM_JOB_NODELIST"
sbatch --nodes=2 --ntasks=384 --time=00:10:00 -A myallocation -p queue-name myjobsubmissionfile.sub
Compute nodes allocated:
gautschi-a[014-015]
The above example will allocate the total of 384 CPU cores across 2 nodes. Note that if your multi-node job requests fewer than each node's full 192 cores per node, by default Slurm provides no guarantee with respect to how this total is distributed between assigned nodes (i.e. the cores may not necessarily be split evenly). If you need specific arrangements of your tasks and cores, you can use --cpus-per-task= and/or --ntasks-per-node= flags. See Slurm documentation or man sbatch for more options.
Directives
So far these examples have shown submitting jobs with the resource requests on the sbatch command line such as:
sbatch -A myallocation -p queue-name --nodes=1 --time=00:01:00 hello.sub
The resource requests can also be put into job submission file itself. Documenting the resource requests in the job submission is desirable because the job can be easily reproduced later. Details left in your command history are quickly lost. Arguments are specified with the #SBATCH syntax:
#!/bin/bash # FILENAME: hello.sub#SBATCH -A myallocation -p queue-name #SBATCH --nodes=1 --time=00:01:00 # Show this ran on a compute node by running the hostname command. hostname echo "Hello World"
The #SBATCH directives must appear at the top of your submission file. SLURM will stop parsing directives as soon as it encounters a line that does not start with '#'. If you insert a directive in the middle of your script, it will be ignored.
This job can be then submitted with:
sbatch hello.sub
Interactive Jobs
Interactive jobs are run on compute nodes, while giving you a shell to interact with. They give you the ability to type commands or use a graphical interface in the same way as if you were on a front-end login host.
To submit an interactive job, use sinteractive to run a login shell on allocated resources.
sinteractive accepts most of the same resource requests as sbatch, so to request a login shell on the cpu account while allocating 2 nodes and 192 total cores, you might do:
sinteractive -A myallocation -p cpu -N2 -n384
To quit your interactive job:
exit or Ctrl-D
The above example will allocate the total of 384 CPU cores across 2 nodes. Note that if your multi-node job requests fewer than each node's full 192 cores per node, by default Slurm provides no guarantee with respect to how this total is distributed between assigned nodes (i.e. the cores may not necessarily be split evenly). If you need specific arrangements of your tasks and cores, you can use --cpus-per-task= and/or --ntasks-per-node= flags. See Slurm documentation or man salloc for more options.
Serial Jobs
This shows how to submit one of the serial programs compiled in the section Compiling Serial Programs.
Create a job submission file:
#!/bin/bash
# FILENAME: serial_hello.sub
./serial_hello
Submit the job:
sbatch --nodes=1 --ntasks=1 --time=00:01:00 serial_hello.sub
After the job completes, view results in the output file:
cat slurm-myjobid.out
Runhost:gautschi-a009.rcac.purdue.edu
hello, world
If the job failed to run, then view error messages in the file slurm-myjobid.out.
Link to section 'Collecting System Resource Utilization Data' of 'Monitoring Resources' Collecting System Resource Utilization Data
Knowing the precise resource utilization an application had during a job, such as CPU load or memory, can be incredibly useful. This is especially the case when the application isn't performing as expected.
One approach is to run a program like htop during an interactive job and keep an eye on system resources. You can get precise time-series data from nodes associated with your job using XDmod as well, online. But these methods don't gather telemetry in an automated fashion, nor do they give you control over the resolution or format of the data.
As a matter of course, a robust implementation of some HPC workload would include resource utilization data as a diagnostic tool in the event of some failure.
The monitor utility is a simple command line system resource monitoring tool for gathering such telemetry and is available as a module.
module load monitor
Complete documentation is available online at resource-monitor.readthedocs.io. A full manual page is also available for reference, man monitor.
In the context of a SLURM job you will need to put this monitoring task in the background to allow the rest of your job script to proceed. Be sure to interrupt these tasks at the end of your job.
#!/bin/bash
# FILENAME: monitored_job.sh
module load monitor
# track per-code CPU load
monitor cpu percent --all-cores >cpu-percent.log &
CPU_PID=$!
# track memory usage
monitor cpu memory >cpu-memory.log &
MEM_PID=$!
# your code here
# shut down the resource monitors
kill -s INT $CPU_PID $MEM_PID
A particularly elegant solution would be to include such tools in your prologue script and have the tear down in your epilogue script.
For large distributed jobs spread across multiple nodes, mpiexec can be used to gather telemetry from all nodes in the job. The hostname is included in each line of output so that data can be grouped as such. A concise way of constructing the needed list of hostnames in SLURM is to simply use srun hostname | sort -u.
#!/bin/bash
# FILENAME: monitored_job.sh
module load monitor
# track all CPUs (one monitor per host)
mpiexec -machinefile <(srun hostname | sort -u) \
monitor cpu percent --all-cores >cpu-percent.log &
CPU_PID=$!
# track memory on all hosts (one monitor per host)
mpiexec -machinefile <(srun hostname | sort -u) \
monitor cpu memory >cpu-memory.log &
MEM_PID=$!
# your code here
# shut down the resource monitors
kill -s INT $CPU_PID $MEM_PID
To get resource data in a more readily computable format, the monitor program can be told to output in CSV format with the --csv flag.
monitor cpu memory --csv >cpu-memory.csv
For a distributed job you will need to suppress the header lines otherwise one will be created by each host.
monitor cpu memory --csv | head -1 >cpu-memory.csv
mpiexec -machinefile <(srun hostname | sort -u) \
monitor cpu memory --csv --no-header >>cpu-memory.csv
Specific Applications
The following examples demonstrate job submission files for some common real-world applications. See the Generic SLURM Examples section for more examples on job submissions that can be adapted for use.
Python
Notice: Python 2.7 has reached end-of-life on Jan 1, 2020 (announcement). Please update your codes and your job scripts to use Python 3.
Python is a high-level, general-purpose, interpreted, dynamic programming language. We suggest using Anaconda which is a Python distribution made for large-scale data processing, predictive analytics, and scientific computing. For example, to use the default Anaconda distribution:
$ module load conda
For a full list of available Anaconda and Python modules enter:
$ module spider conda
Example Python Jobs
This section illustrates how to submit a small Python job to a SLURM queue.
Link to section 'Example 1: Hello world' of 'Example Python Jobs' Example 1: Hello world
Prepare a Python input file with an appropriate filename, here named hello.py:
# FILENAME: hello.py
import string, sys
print("Hello, world!")
Prepare a job submission file with an appropriate filename, here named myjob.sub:
#!/bin/bash
# FILENAME: myjob.sub
module load conda
python hello.py
Hello, world!
Link to section 'Example 2: Matrix multiply' of 'Example Python Jobs' Example 2: Matrix multiply
Save the following script as matrix.py:
# Matrix multiplication program
x = [[3,1,4],[1,5,9],[2,6,5]]
y = [[3,5,8,9],[7,9,3,2],[3,8,4,6]]
result = [[sum(a*b for a,b in zip(x_row,y_col)) for y_col in zip(*y)] for x_row in x]
for r in result:
print(r)
Change the last line in the job submission file above to read:
python matrix.py
The standard output file from this job will result in the following matrix:
[28, 56, 43, 53]
[65, 122, 59, 73]
[63, 104, 54, 60]
Link to section 'Example 3: Sine wave plot using numpy and matplotlib packages' of 'Example Python Jobs' Example 3: Sine wave plot using numpy and matplotlib packages
Save the following script as sine.py:
import numpy as np
import matplotlib
matplotlib.use('Agg')
import matplotlib.pyplot as plt
x = np.linspace(-np.pi, np.pi, 201)
plt.plot(x, np.sin(x))
plt.xlabel('Angle [rad]')
plt.ylabel('sin(x)')
plt.axis('tight')
plt.savefig('sine.png')
Change your job submission file to submit this script and the job will output a png file and blank standard output and error files.
For more information about Python:
Managing Environments with Conda
Conda is a package manager in Anaconda that allows you to create and manage multiple environments where you can pick and choose which packages you want to use. To use Conda you must load an Anaconda module:
$ module load conda
Many packages are pre-installed in the global environment. To see these packages:
$ conda list
To create your own custom environment:
$ conda create --name MyEnvName python=3.8 FirstPackageName SecondPackageName -y
The --name option specifies that the environment created will be named MyEnvName. You can include as many packages as you require separated by a space. Including the -y option lets you skip the prompt to install the package. By default environments are created and stored in the $HOME/.conda directory.
To create an environment at a custom location:
$ conda create --prefix=$HOME/MyEnvName python=3.8 PackageName -y
To see a list of your environments:
$ conda env list
To remove unwanted environments:
$ conda remove --name MyEnvName --all
To add packages to your environment:
$ conda install --name MyEnvName PackageNames
To remove a package from an environment:
$ conda remove --name MyEnvName PackageName
Installing packages when creating your environment, instead of one at a time, will help you avoid dependency issues.
To activate or deactivate an environment you have created:
$ source activate MyEnvName
$ source deactivate MyEnvName
If you created your conda environment at a custom location using --prefix option, then you can activate or deactivate it using the full path.
$ source activate $HOME/MyEnvName
$ source deactivate $HOME/MyEnvName
To use a custom environment inside a job you must load the module and activate the environment inside your job submission script. Add the following lines to your submission script:
$ module load conda
$ source activate MyEnvName
For more information about Python:
Installing Packages
Installing Python packages in an Anaconda environment is recommended. One key advantage of Anaconda is that it allows users to install unrelated packages in separate self-contained environments. Individual packages can later be reinstalled or updated without impacting others. If you are unfamiliar with Conda environments, please check our Conda Guide.
To facilitate the process of creating and using Conda environments, we support a script (conda-env-mod) that generates a module file for an environment, as well as an optional Jupyter kernel to use this environment in a JupyterHub notebook.
You must load one of the anaconda modules in order to use this script.
$ module load conda
Step-by-step instructions for installing custom Python packages are presented below.
Link to section 'Step 1: Create a conda environment' of 'Installing Packages' Step 1: Create a conda environment
Users can use the conda-env-mod script to create an empty conda environment. This script needs either a name or a path for the desired environment. After the environment is created, it generates a module file for using it in future. Please note that conda-env-mod is different from the official conda-env script and supports a limited set of subcommands. Detailed instructions for using conda-env-mod can be found with the command conda-env-mod --help.
-
Example 1: Create a conda environment named mypackages in user's
$HOMEdirectory.$ conda-env-mod create -n mypackages -
Example 2: Create a conda environment named mypackages at a custom location.
$ conda-env-mod create -p /depot/mylab/apps/mypackagesPlease follow the on-screen instructions while the environment is being created. After finishing, the script will print the instructions to use this environment.
... ... ... Preparing transaction: ...working... done Verifying transaction: ...working... done Executing transaction: ...working... done +------------------------------------------------------+ | To use this environment, load the following modules: | | module load use.own | | module load conda-env/mypackages-py3.8.5 | +------------------------------------------------------+ Your environment "mypackages" was created successfully.
Note down the module names, as you will need to load these modules every time you want to use this environment. You may also want to add the module load lines in your jobscript, if it depends on custom Python packages.
By default, module files are generated in your $HOME/privatemodules directory. The location of module files can be customized by specifying the -m /path/to/modules option to conda-env-mod.
Note: The main differences between -p and -m are: 1) -p will change the location of packages to be installed for the env and the module file will still be located at the $HOME/privatemodules directory as defined in use.own. 2) -m will only change the location of the module file. So the method to load modules created with -m and -p are different, see Example 3 for details.
- Example 3: Create a conda environment named labpackages in your group's Data Depot space and place the module file at a shared location for the group to use.
$ conda-env-mod create -p /depot/mylab/apps/labpackages -m /depot/mylab/etc/modules ... ... ... Preparing transaction: ...working... done Verifying transaction: ...working... done Executing transaction: ...working... done +-------------------------------------------------------+ | To use this environment, load the following modules: | | module use /depot/mylab/etc/modules | | module load conda-env/labpackages-py3.8.5 | +-------------------------------------------------------+ Your environment "labpackages" was created successfully.
If you used a custom module file location, you need to run the module use command as printed by the command output above.
By default, only the environment and a module file are created (no Jupyter kernel). If you plan to use your environment in a JupyterHub notebook, you need to append a --jupyter flag to the above commands.
- Example 4: Create a Jupyter-enabled conda environment named labpackages in your group's Data Depot space and place the module file at a shared location for the group to use.
$ conda-env-mod create -p /depot/mylab/apps/labpackages -m /depot/mylab/etc/modules --jupyter ... ... ... Jupyter kernel created: "Python (My labpackages Kernel)" ... ... ... Your environment "labpackages" was created successfully.
Link to section 'Step 2: Load the conda environment' of 'Installing Packages' Step 2: Load the conda environment
-
The following instructions assume that you have used conda-env-mod script to create an environment named mypackages (Examples 1 or 2 above). If you used conda create instead, please use conda activate mypackages.
$ module load use.own $ module load conda-env/mypackages-py3.8.5Note that the conda-env module name includes the Python version that it supports (Python 3.8.5 in this example). This is same as the Python version in the conda module.
-
If you used a custom module file location (Example 3 above), please use module use to load the conda-env module.
$ module use /depot/mylab/etc/modules $ module load conda-env/labpackages-py3.8.5
Link to section 'Step 3: Install packages' of 'Installing Packages' Step 3: Install packages
Now you can install custom packages in the environment using either conda install or pip install.
Link to section 'Installing with conda' of 'Installing Packages' Installing with conda
-
Example 1: Install OpenCV (open-source computer vision library) using conda.
$ conda install opencv -
Example 2: Install a specific version of OpenCV using conda.
$ conda install opencv=4.5.5 -
Example 3: Install OpenCV from a specific anaconda channel.
$ conda install -c anaconda opencv
Link to section 'Installing with pip' of 'Installing Packages' Installing with pip
-
Example 4: Install pandas using pip.
$ pip install pandas -
Example 5: Install a specific version of pandas using pip.
$ pip install pandas==1.4.3Follow the on-screen instructions while the packages are being installed. If installation is successful, please proceed to the next section to test the packages.
Note: Do NOT run Pip with the --user argument, as that will install packages in a different location and might mess up your account environment.
Link to section 'Step 4: Test the installed packages' of 'Installing Packages' Step 4: Test the installed packages
To use the installed Python packages, you must load the module for your conda environment. If you have not loaded the conda-env module, please do so following the instructions at the end of Step 1.
$ module load use.own
$ module load conda-env/mypackages-py3.8.5
- Example 1: Test that OpenCV is available.
$ python -c "import cv2; print(cv2.__version__)" - Example 2: Test that pandas is available.
$ python -c "import pandas; print(pandas.__version__)"
If the commands finished without errors, then the installed packages can be used in your program.
Link to section 'Additional capabilities of conda-env-mod script' of 'Installing Packages' Additional capabilities of conda-env-mod script
The conda-env-mod tool is intended to facilitate creation of a minimal Anaconda environment, matching module file and optionally a Jupyter kernel. Once created, the environment can then be accessed via familiar module load command, tuned and expanded as necessary. Additionally, the script provides several auxiliary functions to help manage environments, module files and Jupyter kernels.
General usage for the tool adheres to the following pattern:
$ conda-env-mod help
$ conda-env-mod <subcommand> <required argument> [optional arguments]
where required arguments are one of
- -n|--name ENV_NAME (name of the environment)
- -p|--prefix ENV_PATH (location of the environment)
and optional arguments further modify behavior for specific actions (e.g. -m to specify alternative location for generated module files).
Given a required name or prefix for an environment, the conda-env-mod script supports the following subcommands:
- create - to create a new environment, its corresponding module file and optional Jupyter kernel.
- delete - to delete existing environment along with its module file and Jupyter kernel.
- module - to generate just the module file for a given existing environment.
- kernel - to generate just the Jupyter kernel for a given existing environment (note that the environment has to be created with a --jupyter option).
- help - to display script usage help.
Using these subcommands, you can iteratively fine-tune your environments, module files and Jupyter kernels, as well as delete and re-create them with ease. Below we cover several commonly occurring scenarios.
Note: When you try to use conda-env-mod delete, remember to include the arguments as you create the environment (i.e. -p package_location and/or -m module_location).
Link to section 'Generating module file for an existing environment' of 'Installing Packages' Generating module file for an existing environment
If you already have an existing configured Anaconda environment and want to generate a module file for it, follow appropriate examples from Step 1 above, but use the module subcommand instead of the create one. E.g.
$ conda-env-mod module -n mypackages
and follow printed instructions on how to load this module. With an optional --jupyter flag, a Jupyter kernel will also be generated.
Note that the module name mypackages should be exactly the same with the older conda environment name. Note also that if you intend to proceed with a Jupyter kernel generation (via the --jupyter flag or a kernel subcommand later), you will have to ensure that your environment has ipython and ipykernel packages installed into it. To avoid this and other related complications, we highly recommend making a fresh environment using a suitable conda-env-mod create .... --jupyter command instead.
Link to section 'Generating Jupyter kernel for an existing environment' of 'Installing Packages' Generating Jupyter kernel for an existing environment
If you already have an existing configured Anaconda environment and want to generate a Jupyter kernel file for it, you can use the kernel subcommand. E.g.
$ conda-env-mod kernel -n mypackages
This will add a "Python (My mypackages Kernel)" item to the dropdown list of available kernels upon your next login to the JupyterHub.
Note that generated Jupiter kernels are always personal (i.e. each user has to make their own, even for shared environments). Note also that you (or the creator of the shared environment) will have to ensure that your environment has ipython and ipykernel packages installed into it.
Link to section 'Managing and using shared Python environments' of 'Installing Packages' Managing and using shared Python environments
Here is a suggested workflow for a common group-shared Anaconda environment with Jupyter capabilities:
The PI or lab software manager:
-
Creates the environment and module file (once):
$ module purge $ module load conda $ conda-env-mod create -p /depot/mylab/apps/labpackages -m /depot/mylab/etc/modules --jupyter -
Installs required Python packages into the environment (as many times as needed):
$ module use /depot/mylab/etc/modules $ module load conda-env/labpackages-py3.8.5 $ conda install ....... # all the necessary packages
Lab members:
-
Lab members can start using the environment in their command line scripts or batch jobs simply by loading the corresponding module:
$ module use /depot/mylab/etc/modules $ module load conda-env/labpackages-py3.8.5 $ python my_data_processing_script.py ..... -
To use the environment in Jupyter notebooks, each lab member will need to create his/her own Jupyter kernel (once). This is because Jupyter kernels are private to individuals, even for shared environments.
$ module use /depot/mylab/etc/modules $ module load conda-env/labpackages-py3.8.5 $ conda-env-mod kernel -p /depot/mylab/apps/labpackages
A similar process can be devised for instructor-provided or individually-managed class software, etc.
Link to section 'Troubleshooting' of 'Installing Packages' Troubleshooting
- Python packages often fail to install or run due to dependency incompatibility with other packages. More specifically, if you previously installed packages in your home directory it is safer to clean those installations.
$ mv ~/.local ~/.local.bak $ mv ~/.cache ~/.cache.bak - Unload all the modules.
$ module purge - Clean up PYTHONPATH.
$ unset PYTHONPATH - Next load the modules (e.g. anaconda) that you need.
$ module load conda/2024.02-py311 $ module load use.own $ module load conda-env/2024.02-py311 - Now try running your code again.
- Few applications only run on specific versions of Python (e.g. Python 3.6). Please check the documentation of your application if that is the case.
Installing Packages from Source
We maintain several Anaconda installations. Anaconda maintains numerous popular scientific Python libraries in a single installation. If you need a Python library not included with normal Python we recommend first checking Anaconda. For a list of modules currently installed in the Anaconda Python distribution:
$ module load conda
$ conda list
# packages in environment at /apps/spack/bell/apps/anaconda/2020.02-py37-gcc-4.8.5-u747gsx:
#
# Name Version Build Channel
_ipyw_jlab_nb_ext_conf 0.1.0 py37_0
_libgcc_mutex 0.1 main
alabaster 0.7.12 py37_0
anaconda 2020.02 py37_0
...
If you see the library in the list, you can simply import it into your Python code after loading the Anaconda module.
If you do not find the package you need, you should be able to install the library in your own Anaconda customization. First try to install it with Conda or Pip. If the package is not available from either Conda or Pip, you may be able to install it from source.
Use the following instructions as a guideline for installing packages from source. Make sure you have a download link to the software (usually it will be a tar.gz archive file). You will substitute it on the wget line below.
We also assume that you have already created an empty conda environment as described in our Python package installation guide.
$ mkdir ~/src
$ cd ~/src
$ wget http://path/to/source/tarball/app-1.0.tar.gz
$ tar xzvf app-1.0.tar.gz
$ cd app-1.0
$ module load conda
$ module load use.own
$ module load conda-env/mypackages-py3.8.5
$ python setup.py install
$ cd ~
$ python
>>> import app
>>> quit()
The "import app" line should return without any output if installed successfully. You can then import the package in your python scripts.
If you need further help or run into any issues installing a library, contact us or drop by Coffee Hour for in-person help.
For more information about Python:
Link to section 'Distributed Training on RCAC Clusters' of 'Distributed Training' Distributed Training on RCAC Clusters
This page aims to provide users of RCAC resources a high-level overview of how distributed training works, along with complete working examples to serve as guides. This guide will cover the following:
- Single node DDP in PyTorch
- Multi-Node DDP in PyTorch
- FSDP in PyTorch
Link to section 'Single Node Distributed Data Parallel' of 'Distributed Training' Single Node Distributed Data Parallel
We will first focus on single node example of Distributed Data Parallelism (DDP) using 8 GPUs for an example mnist classification task. DDP aims to speed up training by training the model on multiple GPUs in parallel. Each instance of the model will receive its own batch of data, and compute its own gradients.
Importantly, before the update step, the gradients across each instance of the model will be accumulated and averaged. This prevents the weights of each of the models from diverging over time.
In this example, we will use torchrun to spawn 8 processes on the node (one for each GPU). Each GPU will receive its own copy of the model, along with its own batch from the dataloader. Pytorch provides us with a comprehensive set of tooling to orchestrate the necessary environment and communications between all of the different processes. We can visualize the setup below. Each process (rank) on the node has its own GPU, with a complete copy of the model.
Link to section 'Slurm Script' of 'Distributed Training' Slurm Script
In the Slurm script, we are requesting a single Gautschi H node (with all 8 H100 GPUs) with a single task. Although we are only requesting a single task, 8 separate processes will be spawned by this task for distributed training.
Before we start training, we must specify the master port and address that will be used for rendezvous communications. This is where all of the individual processes sync up for communication. The Hostname will be formatted like h012.gautschi.rcac.purdue.edu, since this job landed on the h012 node.
In the last line, torchrun is ran only once, and is responsible for managing the spawning of processes (one for each GPU on this node), as well as setting environment variables that will be used within each process for establishing communication:
RANK, LOCAL_RANK, WORLD_SIZE, MASTER_ADDR, MASTER_PORT
Here, the rank (and local rank, which is more relevant for multi-node training, see below), world size, and rendezvous backend and location are all set.
#!/bin/bash
#SBATCH --job-name=ddp_singlenode
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --gpus-per-node=8
#SBATCH --cpus-per-task=14
#SBATCH --time=00:20:00
#SBATCH --constraint=H
#SBATCH --partition=ai
#SBATCH --qos=normal
#SBATCH --output=train_gautschi_singlenode.out
#SBATCH --account=rcac #Change to your account!
#Change these to match your environment!
ml conda
ml cuda
conda activate /depot/itap/user/envs/anaconda/gautschi/ddp_example
# Optional Logging (Uncomment these if you are having issues!)
# export NCCL_DEBUG=INFO
# export NCCL_DEBUG_SUBSYS=ALL
# export TORCH_CPP_LOG_LEVEL=INFO
# export TORCH_DISTRIBUTED_DEBUG=DETAIL
# Rendezvous settings (do this before so torchrun sees them)
# Will be used for establishing the rendezvous point for all processes
export MASTER_ADDR=$(hostname)
export MASTER_PORT=29500
echo "MASTER_ADDR: $MASTER_ADDR" #h012.gautschi.rcac.purdue.edu
echo "MASTER_PORT: $MASTER_PORT"
echo "SLURM_GPUS_ON_NODE: $SLURM_GPUS_PER_NODE" #8
torchrun --nproc_per_node=${SLURM_GPUS_PER_NODE} \
--rdzv_endpoint=${MASTER_ADDR}:${MASTER_PORT} \
ddp_train_gautschi.py
Link to section 'PyTorch Script' of 'Distributed Training' PyTorch Script
The Pytorch script largely looks like a standard Pytorch training script, but with modifications to allow for distributed training. With respect to DDP, the important portions of this script are as follows:
- The
dist.init_process_group(backend="nccl")initializes the distributed process group. This allows for the collective communication across processes. Using environment variables set for each process by torchrun, it connects all processes to a rendezvous endpoint to allow for collective communication before training begins. Effectively, it ensures that all processes understand what their rank is, what all the other processes are, and that all processes are able to communicate with each other. This then allows each process to see information specific to it (such asget_rank()). backend="nccl"specifies the communication backend (Nvidia Collective Communications Library) for the sharing of gradients between processes.- The
sampler = DistributedSampler(train_dataset, num_replicas=world_size, rank=rank)creates a sampler that will send different batches of your training dataset to each of the ranks (GPUs), which each contain their own copy of the model. - The
ddp_model = DDP(model, device_ids=[local_rank])is arguably the most important portion of the code. When you wrap a pytorch model inDDP()it transforms the model into a distributed model with all other ranks. This model will then share weighs with all other distributed models in all of the other ranks (using the nccl backend).
# (filename is ddp_train_gautschi.py)
import os
import torch
import torch.nn as nn
import torch.optim as optim
import torch.distributed as dist
from torch.nn.parallel import DistributedDataParallel as DDP
from torch.utils.data import DataLoader, DistributedSampler
from torchvision import datasets, transforms
import torch.nn.functional as F
import socket
def setup():
dist.init_process_group(backend="nccl")
def cleanup():
dist.destroy_process_group()
class MNISTModel(nn.Module):
'''This is a vanilla CNN compatible with the default mnist dataset!'''
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1, 32, kernel_size=3, padding=1)
self.conv2 = nn.Conv2d(32, 64, kernel_size=3, padding=1)
self.pool = nn.MaxPool2d(2)
self.fc1 = nn.Linear(64 * 7 * 7, 128)
self.fc2 = nn.Linear(128, 10)
def forward(self, x):
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
x = x.view(x.size(0), -1)
x = F.relu(self.fc1(x))
return self.fc2(x)
def train():
setup()
rank = dist.get_rank() #Will be the same as global rank for single-node!
world_size = dist.get_world_size() #Will be the #nodes * gpus/node!
local_rank = int(os.environ["LOCAL_RANK"]) #LOCAL_RANK managed by torchrun!
torch.cuda.set_device(local_rank) #LOCAL_RANK is just the GPU index to set!
device = torch.device("cuda", local_rank)
print(f"Rank:{rank}, local_rank:{local_rank}, hostname:{socket.gethostname()}, world_size = {world_size}", flush=True)
#Load standard mnist dataset !
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))
])
train_dataset = datasets.MNIST(root="./data", train=True, download=True, transform=transform)
#DistributedSampler will give each rank (gpu) its own batch!
sampler = DistributedSampler(train_dataset, num_replicas=world_size, rank=rank)
dataloader = DataLoader(train_dataset, batch_size=64, sampler=sampler, num_workers=4, pin_memory=True)
#Wrapping model in DDP allows for the sync of gradients across all ranks (gpus)!
model = MNISTModel().to(device)
ddp_model = DDP(model, device_ids=[local_rank])
optimizer = optim.Adam(ddp_model.parameters(), lr=0.001)
loss_fn = nn.CrossEntropyLoss()
#After that the "training loop" is basically identical to a
for epoch in range(5):
ddp_model.train()
sampler.set_epoch(epoch)
total_loss = 0.0
for batch_x, batch_y in dataloader:
batch_x = batch_x.to(device, non_blocking=True)
batch_y = batch_y.to(device, non_blocking=True)
optimizer.zero_grad()
output = ddp_model(batch_x)
loss = loss_fn(output, batch_y)
loss.backward()
optimizer.step()
total_loss += loss.item()
print(f"Rank {rank}, Epoch {epoch},Loss: {total_loss / len(dataloader):.4f}", flush=True)
print("done training", flush=True)
# Only save the model once (from the first rank!)
if rank == 0:
torch.save(ddp_model.state_dict(), "mnist_ddp_model.pth")
print("Model saved", flush=True)
cleanup()
if __name__ == "__main__":
train()
Link to section 'Output' of 'Distributed Training' Output
From the output, we see that the Python script is running 8 different times, with 8 separate ranks:
Rank:0, local_rank:0, hostname:h007.gautschi.rcac.purdue.edu, world_size = 8
Rank:2, local_rank:2, hostname:h007.gautschi.rcac.purdue.edu, world_size = 8
Rank:4, local_rank:4, hostname:h007.gautschi.rcac.purdue.edu, world_size = 8
Rank:3, local_rank:3, hostname:h007.gautschi.rcac.purdue.edu, world_size = 8
Rank:1, local_rank:1, hostname:h007.gautschi.rcac.purdue.edu, world_size = 8
Rank:7, local_rank:7, hostname:h007.gautschi.rcac.purdue.edu, world_size = 8
Rank:5, local_rank:5, hostname:h007.gautschi.rcac.purdue.edu, world_size = 8
Rank:6, local_rank:6, hostname:h007.gautschi.rcac.purdue.edu, world_size = 8
Link to section 'Multi Node Distributed Data Parallel' of 'Distributed Training' Multi Node Distributed Data Parallel
Things become a bit more complicated when we want to distribute training across multiple nodes. With a total world size of 16 (2 nodes, with 8 ranks each) each process will have a "global rank" (1-16) as well as a "local rank" on that node, which will correspond the the GPU ID:
Link to section 'Slurm Script' of 'Distributed Training' Slurm Script
In this Slurm script, we are now requesting two Gautschi H nodes (each with all 8 H100 GPUs), and specify that each node should have a single task. Again, the single task on each node will spawn 8 separate processes (corresponding to the 8 local ranks) per node.
Before we start training, we must specify the master port and address that will be used for rendezvous communications. This is where all of the individual processes sync up for communication. Since there are now multiple nodes, we must pull out the first node (from $SLURM_JOB_NODELIST) to serve as the master.
When we run srun torchrun here, and since we requested 1 task per node, torchrun is being ran once on each node, which in-turn spawns 8 processes (one for each rank) on each node.
#!/bin/bash
#SBATCH --job-name=ddp_multinode
#SBATCH --nodes=2
#SBATCH --ntasks-per-node=1
#SBATCH --gpus-per-node=8
#SBATCH --cpus-per-task=14
#SBATCH --time=00:05:00
#SBATCH --constraint=H
#SBATCH --partition=ai
#SBATCH --qos=normal
#SBATCH --output=train_gautschi_multinode.out
#SBATCH --account=rcac
ml conda
ml cuda
conda activate /depot/itap/user/envs/anaconda/gautschi/ddp_example
export MASTER_ADDR=$(scontrol show hostnames "$SLURM_JOB_NODELIST" | head -n1)
export MASTER_PORT=29500
srun torchrun \
--nnodes=${SLURM_NNODES} \
--node_rank=${SLURM_NODEID} \
--nproc_per_node=${SLURM_GPUS_PER_NODE} \
--rdzv_id=${SLURM_JOB_ID} \
--rdzv_backend=c10d \
--rdzv_endpoint=${MASTER_ADDR}:${MASTER_PORT} \
ddp_train_gautschi.py
Link to section 'PyTorch Script' of 'Distributed Training' PyTorch Script
Torchrun is handling all of the processes and environment variables that are needed to initiate the distributed process group, so no changes to the python script are needed!
Link to section 'Output' of 'Distributed Training' Output
From the output, we see that the Python script is now running 16 different times, with 8 processes on each node!
Rank:0, local_rank:0, hostname:h014.gautschi.rcac.purdue.edu, world_size = 16
Rank:8, local_rank:0, hostname:h016.gautschi.rcac.purdue.edu, world_size = 16
Rank:1, local_rank:1, hostname:h014.gautschi.rcac.purdue.edu, world_size = 16
Rank:3, local_rank:3, hostname:h014.gautschi.rcac.purdue.edu, world_size = 16
Rank:6, local_rank:6, hostname:h014.gautschi.rcac.purdue.edu, world_size = 16
Rank:4, local_rank:4, hostname:h014.gautschi.rcac.purdue.edu, world_size = 16
Rank:2, local_rank:2, hostname:h014.gautschi.rcac.purdue.edu, world_size = 16
Rank:7, local_rank:7, hostname:h014.gautschi.rcac.purdue.edu, world_size = 16
Rank:5, local_rank:5, hostname:h014.gautschi.rcac.purdue.edu, world_size = 16
Rank:9, local_rank:1, hostname:h016.gautschi.rcac.purdue.edu, world_size = 16
Rank:10, local_rank:2, hostname:h016.gautschi.rcac.purdue.edu, world_size = 16
Rank:15, local_rank:7, hostname:h016.gautschi.rcac.purdue.edu, world_size = 16
Rank:14, local_rank:6, hostname:h016.gautschi.rcac.purdue.edu, world_size = 16
Rank:11, local_rank:3, hostname:h016.gautschi.rcac.purdue.edu, world_size = 16
Rank:12, local_rank:4, hostname:h016.gautschi.rcac.purdue.edu, world_size = 16
Rank:13, local_rank:5, hostname:h016.gautschi.rcac.purdue.edu, world_size = 16
Link to section 'Fully Sharded Data Parallel' of 'Distributed Training' Fully Sharded Data Parallel
Fully Sharded Data Parallel (FSDP) is useful if your model is too large to fit on a single GPU. FSDP allows you to shard your model across multiple GPUs, and lets NCCL handle communincation between them. The Slurm submission script looks almost identical to our single-node DDP example above:
#!/bin/bash
#SBATCH --job-name=fsdp_singlenode
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --gpus-per-node=8
#SBATCH --cpus-per-task=14
#SBATCH --time=00:20:00
#SBATCH --constraint=H
#SBATCH --partition=ai
#SBATCH --qos=normal
#SBATCH --output=fsdp_train_gautschi_singlenode.out
#SBATCH --account=rcac
#Change these to match your environment!
ml conda
ml cuda
conda activate /depot/itap/verburgt/envs/anaconda/gautschi/ddp_example
# Optional Logging (Uncomment these if you are having issues!)
# export NCCL_DEBUG=INFO
# export NCCL_DEBUG_SUBSYS=ALL
# export TORCH_CPP_LOG_LEVEL=INFO
# export TORCH_DISTRIBUTED_DEBUG=DETAIL
# Rendezvous settings (do this before so torchrun sees them)
# Will be used for establishing the rendezvous point for all processes
export MASTER_ADDR=$(hostname)
export MASTER_PORT=29500
echo "MASTER_ADDR: $MASTER_ADDR"
echo "MASTER_PORT: $MASTER_PORT"
echo "SLURM_GPUS_ON_NODE: $SLURM_GPUS_PER_NODE"
torchrun --nproc_per_node=${SLURM_GPUS_PER_NODE} \
--rdzv_endpoint=${MASTER_ADDR}:${MASTER_PORT} \
fsdp_train_gautschi.py
Changes are primarily made to the python script:
import os
import torch
import torch.nn as nn
import torch.optim as optim
import torch.distributed as dist
from torch.distributed.fsdp import FullyShardedDataParallel as FSDP
from torch.distributed.fsdp import StateDictType, FullStateDictConfig, ShardedStateDictConfig
from torch.distributed.fsdp.wrap import size_based_auto_wrap_policy
from torch.distributed.fsdp import ShardingStrategy
from torch.utils.data import DataLoader
from torch.utils.data.distributed import DistributedSampler
from torchvision import datasets, transforms
import torch.nn.functional as F
import socket
from functools import partial
def setup():
dist.init_process_group(backend="nccl")
def cleanup():
dist.destroy_process_group()
class MNISTModel(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(1, 32, kernel_size=3, padding=1)
self.conv2 = nn.Conv2d(32, 64, kernel_size=3, padding=1)
self.pool = nn.MaxPool2d(2)
self.fc1 = nn.Linear(64 * 7 * 7, 128)
self.fc2 = nn.Linear(128, 10)
def forward(self, x):
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
x = x.view(x.size(0), -1)
x = F.relu(self.fc1(x))
return self.fc2(x)
def train():
setup()
rank = dist.get_rank()
world_size = dist.get_world_size()
local_rank = int(os.environ["LOCAL_RANK"])
torch.cuda.set_device(local_rank)
device = torch.device("cuda", local_rank)
print(f"Rank:{rank}, local_rank:{local_rank}, hostname:{socket.gethostname()}, world_size = {world_size}", flush=True)
# Transform and dataset
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))
])
train_dataset = datasets.MNIST(root="./data", train=True, download=True, transform=transform)
sampler = DistributedSampler(train_dataset, num_replicas=world_size, rank=rank)
dataloader = DataLoader(train_dataset, batch_size=64, sampler=sampler, num_workers=4, pin_memory=True)
# Initialize FSDP model
# We define a wrap policy to allow our relatively small model to be fully sharded
base_model = MNISTModel().to(device)
wrap_policy = partial(size_based_auto_wrap_policy, min_num_params=1)
# Create the FSDP Model with the wrap policy
fsdp_model = FSDP(
base_model,
auto_wrap_policy=wrap_policy,
sharding_strategy=ShardingStrategy.FULL_SHARD,
)
#Show the number of parameters in each rank (for demonstration purposes)
param_count = sum(p.numel() for p in base_model.parameters()) #The un-sharded model
local_cnt = sum(p.numel() for p in fsdp_model.parameters()) # The sharded model
t = torch.tensor([local_cnt], device=device)
gathered = [torch.zeros_like(t) for _ in range(world_size)]
dist.all_gather(gathered, t)
if rank == 0:
cnts = [int(x.item()) for x in gathered]
print("Local param counts per rank:", cnts) #There might be extra padding in some shards!
print("Total parameters:", param_count)
optimizer = optim.Adam(fsdp_model.parameters(), lr=0.001)
loss_fn = nn.CrossEntropyLoss()
for epoch in range(5):
sampler.set_epoch(epoch)
fsdp_model.train()
total_loss = 0.0
for batch_x, batch_y in dataloader:
batch_x = batch_x.to(device, non_blocking=True)
batch_y = batch_y.to(device, non_blocking=True)
optimizer.zero_grad()
output = fsdp_model(batch_x)
loss = loss_fn(output, batch_y)
loss.backward()
optimizer.step()
total_loss += loss.item()
print(f"Rank {rank}, Epoch {epoch}, Avg Loss: {total_loss / len(dataloader):.4f}", flush=True)
print("done training", flush=True)
#Aggregate the model and save it from rank 0
dist.barrier() #Ensure all ranks are done
# Build a full state dict but only on rank 0
full_cfg = FullStateDictConfig(offload_to_cpu=True, rank0_only=True)
with FSDP.state_dict_type(fsdp_model, StateDictType.FULL_STATE_DICT, full_cfg):
state_dict = fsdp_model.state_dict()
#Save full model from only rank 0
if rank == 0:
torch.save(state_dict, "mnist_fsdp_full.pth")
print("Model saved", flush=True)
# Cam also save shards from each rank
with FSDP.state_dict_type(fsdp_model, StateDictType.SHARDED_STATE_DICT, ShardedStateDictConfig()):
shard = fsdp_model.state_dict()
torch.save(shard, f"mnist_fsdp_shard_rank{rank}.pth")
dist.barrier()
cleanup()
if __name__ == "__main__":
train()
The primary differences here are in saving the model. Instead of using DDP to create the model, we instead use FSDP, along with a "wrap policy", which defines how, and which parts of our model, are to be wrapped inside FSDP. We also save the model differently. Since the model is sharded across multiple GPUs, we must aggregate the model onto a single node before saving the complete model. Alternatively, we can save the sharded model from each rank individually.
Link to section 'Investigating inter-process communications' of 'Distributed Training' Investigating inter-process communications
If you enabled detailed logging by uncommenting these variables:
export NCCL_DEBUG=INFO
export NCCL_DEBUG_SUBSYS=ALL
export TORCH_CPP_LOG_LEVEL=INFO
export TORCH_DISTRIBUTED_DEBUG=DETAIL
You will be able to see detailed information reported by the rendezvous and NCCL backends. (**WARNING** This will produce *very* large logs)
When doing gradient updating, you will see that NCCL is handling the synchronization of data between ranks (with the AllReduce function):
h015:2073494:2073958 [2] NCCL INFO AllReduce: opCount 1e7 sendbuff 0x145d0be7e400 recvbuff 0x145d0be7e400 count 18816 datatype 7 op 0 root 0 comm 0x55956223bb90 [nranks=16] stream 0x55956223a9a0
You can also see that each of the ranks are frequently accessing the rendezvous backend (c10d) for getting and setting information about other ranks:
[I521 16:05:33.213697937 TCPStoreLibUvBackend.cpp:827] [c10d - trace] set key:/default_pg/0//cuda//NCCL_4_trace_start address:[h005.gautschi.rcac.purdue.edu]:57348
Link to section 'FAQ' of 'Distributed Training' FAQ
- Is the
NCCL"process group" backend different than the "rendezvous" backend? What's the difference?- The rendezvous backend (typically
c10d) is what is used to allow different ranks to find, discover, and communicate with each other. Under the hood, it's effectively just a TCPStore key-value server that all ranks have access to. In the logs, you'll see each rank is often getting and setting values, which will be visible to all other ranks.
- The rendezvous backend (typically
* The NCCL (Nvidia Collective Communications Library) process group backend is what is facilitating distributed operations of for GPUs. For example, NCCL is handling the AllReduce function, which is averaging gradients across GPUs when backpropgating in a DDP() model. There are other process group backends (gloo, mpi) but nccl is strongly recommended for use with Nvidia GPUs.
- Do I need to use
torchrun? What exactly istorchrundoing?- You don't necessarily need to use
torchrun, and could launch all processes withsrun, or even manually. However, you'll need to ensure that the environment for each rank are set up properly, which would be tedious and error-prone without torchrun.
- You don't necessarily need to use
- How can I ensure all of the individual processes are synced?
- In the
setup()function, you can add atorch.distributed.barrier()line, which will prevent all processes from continuing until all have reached the barrier.
- In the
- Can I run distributed training in a container?
- Yes! You can use NGC (Nvidia GPU Cloud) containers to perform distributed training. Running with singularity may look something like this:
singularity run --nv /apps/ngc/images/nvcr.io_nvidia_pytorch:25.01-py3.sif \
torchrun --nproc_per_node=$SLURM_GPUS_PER_NODE \
--rdzv-backend=c10d \
--rdzv-endpoint=$MASTER_ADDR:$MASTER_PORT \
ddp_train_gautschi.py
R
R, a GNU project, is a language and environment for data manipulation, statistics, and graphics. It is an open source version of the S programming language. R is quickly becoming the language of choice for data science due to the ease with which it can produce high quality plots and data visualizations. It is a versatile platform with a large, growing community and collection of packages.
For more general information on R visit The R Project for Statistical Computing.
Loading Data into R
R is an environment for manipulating data. In order to manipulate data, it must be brought into the R environment. R has a function to read any file that data is stored in. Some of the most common file types like comma-separated variable(CSV) files have functions that come in the basic R packages. Other less common file types require additional packages to be installed. To read data from a CSV file into the R environment, enter the following command in the R prompt:
> read.csv(file = "path/to/data.csv", header = TRUE)
When R reads the file it creates an object that can then become the target of other functions. By default the read.csv() function will give the object the name of the .csv file. To assign a different name to the object created by read.csv enter the following in the R prompt:
> my_variable <- read.csv(file = "path/to/data.csv", header = FALSE)
To display the properties (structure) of loaded data, enter the following:
> str(my_variable)
For more functions and tutorials:
Installing R packages
Link to section 'Challenges of Managing R Packages in the Cluster Environment' of 'Installing R packages' Challenges of Managing R Packages in the Cluster Environment
- Different clusters have different hardware and softwares. So, if you have access to multiple clusters, you must install your R packages separately for each cluster.
- Each cluster has multiple versions of R and packages installed with one version of R may not work with another version of R. So, libraries for each R version must be installed in a separate directory.
- You can define the directory where your R packages will be installed using the environment variable
R_LIBS_USER. - For your convenience, a sample ~/.Rprofile example file is provided that can be downloaded to your cluster account and renamed into
~/.Rprofile(or appended to one) to customize your installation preferences. Detailed instructions.
Link to section 'Installing Packages' of 'Installing R packages' Installing Packages
-
Step 0: Set up installation preferences.
Follow the steps for setting up your~/.Rprofilepreferences. This step needs to be done only once. If you have created a~/.Rprofilefile previously on Gautschi, ignore this step. -
Step 1: Check if the package is already installed.
As part of the R installations on community clusters, a lot of R libraries are pre-installed. You can check if your package is already installed by opening an R terminal and entering the commandinstalled.packages(). For example,module load r/4.4.1 Rinstalled.packages()["units",c("Package","Version")] Package Version "units" "0.8-1" quit()If the package you are trying to use is already installed, simply load the library, e.g.,
library('units'). Otherwise, move to the next step to install the package. -
Step 2: Load required dependencies. (if needed)
For simple packages you may not need this step. However, some R packages depend on other libraries. For example, thesfpackage depends ongdalandgeoslibraries. So, you will need to load the corresponding modules before installingsf. Read the documentation for the package to identify which modules should be loaded.module load gdal module load geos -
Step 3: Install the package.
Now install the desired package using the commandinstall.packages('package_name'). R will automatically download the package and all its dependencies from CRAN and install each one. Your terminal will show the build progress and eventually show whether the package was installed successfully or not.Rinstall.packages('sf', repos="https://cran.case.edu/") Installing package into ‘/home/myusername/R/x86_64-pc-linux-gnu-library/4.4.1’ (as ‘lib’ is unspecified) trying URL 'https://cran.case.edu/src/contrib/sf_0.9-7.tar.gz' Content type 'application/x-gzip' length 4203095 bytes (4.0 MB) ================================================== downloaded 4.0 MB ... ... more progress messages ... ... ** testing if installed package can be loaded from final location ** testing if installed package keeps a record of temporary installation path * DONE (sf) The downloaded source packages are in ‘/tmp/RtmpSVAGio/downloaded_packages’ - Step 4: Troubleshooting. (if needed)
If Step 3 ended with an error, you need to investigate why the build failed. Most common reason for build failure is not loading the necessary modules.
Link to section 'Loading Libraries' of 'Installing R packages' Loading Libraries
Once you have packages installed you can load them with the library() function as shown below:
library('packagename')
The package is now installed and loaded and ready to be used in R.
Link to section 'Example: Installing dplyr' of 'Installing R packages' Example: Installing dplyr
The following demonstrates installing the dplyr package assuming the above-mentioned custom ~/.Rprofile is in place (note its effect in the "Installing package into" information message):
module load r
R
install.packages('dplyr', repos="http://ftp.ussg.iu.edu/CRAN/")
Installing package into ‘/home/myusername/R/gautschi/4.4.1’
(as ‘lib’ is unspecified)
...
also installing the dependencies 'crayon', 'utf8', 'bindr', 'cli', 'pillar', 'assertthat', 'bindrcpp', 'glue', 'pkgconfig', 'rlang', 'Rcpp', 'tibble', 'BH', 'plogr'
...
...
...
The downloaded source packages are in
'/tmp/RtmpHMzm9z/downloaded_packages'
library(dplyr)
Attaching package: 'dplyr'
For more information about installing R packages:
RStudio
RStudio is a graphical integrated development environment (IDE) for R. RStudio is the most popular environment for developing both R scripts and packages. RStudio is provided on most Research systems.
There are two methods to launch RStudio on the cluster: command-line and application menu icon.
Link to section 'Launch RStudio by the command-line:' of 'RStudio' Launch RStudio by the command-line:
module load gcc
module load r
module load rstudio
rstudio
Note that RStudio is a graphical program and in order to run it you must have a local X11 server running or use Thinlinc Remote Desktop environment. See the ssh X11 forwarding section for more details.
Link to section 'Launch Rstudio by the application menu icon:' of 'RStudio' Launch Rstudio by the application menu icon:
- Log into desktop.gautschi.rcac.purdue.edu with web browser or ThinLinc client
- Click on the
Applicationsdrop down menu on the top left corner - Choose
Cluster Softwareand thenRStudio
![]()
R and RStudio are free to download and run on your local machine. For more information about RStudio:
Running R jobs
This section illustrates how to submit a small R job to a SLURM queue. The example job computes a Pythagorean triple.
Prepare an R input file with an appropriate filename, here named myjob.R:
# FILENAME: myjob.R
# Compute a Pythagorean triple.
a = 3
b = 4
c = sqrt(a*a + b*b)
c # display result
Prepare a job submission file with an appropriate filename, here named myjob.sub:
#!/bin/bash
# FILENAME: myjob.sub
module load r
# --vanilla:
# --no-save: do not save datasets at the end of an R session
R --vanilla --no-save < myjob.R
For other examples or R jobs:
Singularity
On Gautschi, Singularity functionality is provided by Apptainer - see Apptainer section for details.
Apptainer
Note: Apptainer was formerly known as Singularity and is now a part of the Linux Foundation. When migrating from Singularity see the user compatibility documentation.
Link to section 'What is Apptainer?' of 'Apptainer' What is Apptainer?
Apptainer is an open-source container platform designed to be simple, fast, and secure. It allows the portability and reproducibility of operating systems and application environments through the use of Linux containers. It gives users complete control over their environment.
Apptainer is like Docker but tuned explicitly for HPC clusters. More information is available on the project’s website.
Link to section 'Features' of 'Apptainer' Features
- Run the latest applications on an Ubuntu or Centos userland
- Gain access to the latest developer tools
- Launch MPI programs easily
- Much more
Apptainer’s user guide is available at: apptainer.org/docs/user/main/introduction.html
Link to section 'Example' of 'Apptainer' Example
Here is an example using an Ubuntu 16.04 image on Gautschi:
apptainer exec /depot/itap/singularity/ubuntu1604.img cat /etc/lsb-release
DISTRIB_ID=Ubuntu
DISTRIB_RELEASE=16.04
DISTRIB_CODENAME=xenial
DISTRIB_DESCRIPTION="Ubuntu 16.04 LTS"
Here is another example using a Centos 7 image:
apptainer exec /depot/itap/singularity/centos7.img cat /etc/redhat-release
CentOS Linux release 7.3.1611 (Core)
Link to section 'Purdue Cluster Specific Notes' of 'Apptainer' Purdue Cluster Specific Notes
All service providers will integrate Apptainer slightly differently depending on site. The largest customization will be which default files are inserted into your images so that routine services will work.
Services we configure for your images include DNS settings and account information. File systems we overlay into your images are your home directory, scratch, Data Depot, and application file systems.
Here is a list of paths:
- /etc/resolv.conf
- /etc/hosts
- /home/$USER
- /apps
- /scratch
- /depot
This means that within the container environment these paths will be present and the same as outside the container. The /apps, /scratch, and /depot directories will need to exist inside your container to work properly.
Link to section 'Creating Apptainer Images' of 'Apptainer' Creating Apptainer Images
You can build on your system or straight on the cluster (you do not need root privileges to build or run the container).
You can find information and documentation for how to install and use Apptainer on your system:
We have version 1.1.6 (or newer) on the cluster. Please note that installed versions may change throughout cluster life time, so when in doubt, please check exact version with a --version command line flag:
apptainer --version
apptainer version 1.3.3-1.el9
Everything you need on how to build a container is available from their user guide. Below are merely some quick tips for getting your own containers built for Gautschi.
You can use a Definition File to both build your container and share its specification with collaborators (for the sake of reproducibility). Here is a simplistic example of such a file:
# FILENAME: Buildfile
Bootstrap: docker
From: ubuntu:18.04
%post
apt-get update && apt-get upgrade -y
mkdir /apps /depot /scratch
To build the image itself:
apptainer build ubuntu-18.04.sif Buildfile
The challenge with this approach however is that it must start from scratch if you decide to change something. In order to create a container image iteratively and interactively, you can use the --sandbox option.
apptainer build --sandbox ubuntu-18.04 docker://ubuntu:18.04
This will not create a flat image file but a directory tree (i.e., a folder), the contents of which are the container's filesystem. In order to get a shell inside the container that allows you to modify it, user the --writable option.
apptainer shell --writable ubuntu-18.04
Apptainer>
You can then proceed to install any libraries, software, etc. within the container. Then to create the final image file, exit the shell and call the build command once more on the sandbox.
apptainer build ubuntu-18.04.sif ubuntu-18.04
Finally, copy the new image to Gautschi and run it.
GPU Usage Monitoring
Link to section 'What is it?' of 'GPU Usage Monitoring' What is it?
To ensure that GPUs are effectively utilized on our cluster, we log and store the power, memory, and utilization of every GPU on our cluster every 10 seconds. RCAC uses this data to identify users that request large amounts of GPU hours but do not actually use them.
To assist users in optimizing their usage of requested GPU resources, we have created a GPU usage monitor tool that allows users to track their GPU utilization for jobs that they have submitted. The purpose of this tool is to provide users an interface to identify jobs or GPUs that they have requested but are not being effectively utilized.
Link to section 'How to use' of 'GPU Usage Monitoring' How to use
Our GPU Usage monitor is currently implemented on Gautschi and Gilbreth, and is available at the following location:
/apps/external/gpu_util/get_gpu_util
Link to section 'Seeing Logged Jobs' of 'GPU Usage Monitoring' Seeing Logged Jobs
You can check all the jobs that you've run with the -u or --user_jobs flag:
/apps/external/gpu_util/get_gpu_util --user_jobs
This will print a table of all the GPU jobs you have submitted to the cluster, with the job ID, start and end times, and the number of GPUs that that job had allocated to it:
It should be noted that we collect GPU usage information every 3 - 3.5 hours. If you have recently submitted a job, your job may not be available yet.
Link to section 'Inspecting a specific job' of 'GPU Usage Monitoring' Inspecting a specific job
If you want to view the GPU utilization of a specific job, you can use the -j or --job_id flag. For example, to see the utilization of job 32083, you may run the following:
/apps/external/gpu_util/get_gpu_util --job_id 32083
This will print a table of the GPU memory and utilization for each GPU that was allocated for the specified job:
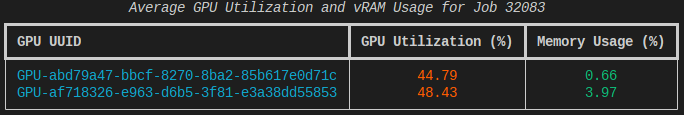
Link to section 'Plotting Data' of 'GPU Usage Monitoring' Plotting Data
It is also possible to plot the GPU utilization and memory usage over time with the -p or the --plot flag:
/apps/external/gpu_util/get_gpu_util --job_id 32083 --plot
This will print two graphs. The first will be a 2-minute rolling average of the GPU utilization over time, and the second will be the percentage of memory used over time. Each GPU will be colored differently:
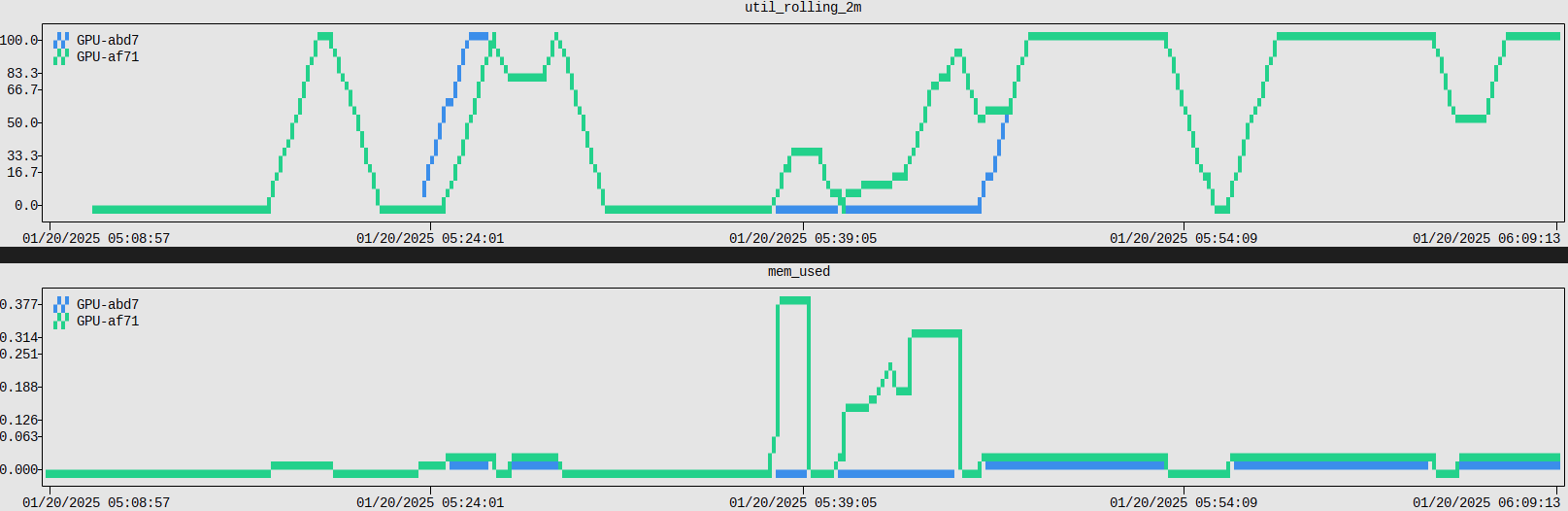
Link to section 'Saving GPU Data' of 'GPU Usage Monitoring' Saving GPU Data
If you would like to save the raw GPU utilization and memory usage data for your job, you can use the -s or --save flag:
/apps/external/gpu_util/get_gpu_util --job_id 32083 --save
This will download the GPU data for a specific job as:
{job_id}_gpu_usage.csv
Link to section 'Checking Account Usage (Group Managers only)' of 'GPU Usage Monitoring' Checking Account Usage (Group Managers only)
If you are a manager of a specific group, you are able to query all accounts owned by that group. You can check the usage of all members of your account over the previous week by running:
/apps/external/gpu_util/get_gpu_util -A accountnameIf you would like a breakdown of the individual jobs ran within the past week, you can add the --records flag:
/apps/external/gpu_util/get_gpu_util -A accountname --recordsFurther, the --job {jobid} flag will also allow you to query the utilization of individual jobs that were ran on an account that you manage.
Link to section 'FAQ/Troubleshooting' of 'GPU Usage Monitoring' FAQ/Troubleshooting
- "Why doesn't my job show up in the list?"
- Although we log GPU utilization every 10 seconds, we only actually collect it every 3-3.5 hours. If you check later, your job should be listed.
- "Can other people see my data?"
- No, our interface only allows users to check the utilization of their own jobs.
- "Is this a good way to see the GPU Hours balance?"
- No, the primary purpose of this tool is to check that GPUs are actually being used, not to track billed GPU hours. To see the total GPU hours balance, please use the
slistcommand.
- No, the primary purpose of this tool is to check that GPUs are actually being used, not to track billed GPU hours. To see the total GPU hours balance, please use the
File Storage and Transfer
Learn more about file storage transfer for Gautschi.
Storage Options
File storage options on RCAC systems include long-term storage (home directories, depot, Fortress) and short-term storage (scratch directories, /tmp directory). Each option has different performance and intended uses, and some options vary from system to system as well. Daily snapshots of home directories are provided for a limited time for accidental deletion recovery. Scratch directories and temporary storage are not backed up and old files are regularly purged from scratch and /tmp directories. More details about each storage option appear below.
Home Directory
Home directories are provided for long-term file storage. Each user has one home directory. You should use your home directory for storing important program files, scripts, input data sets, critical results, and frequently used files. You should store infrequently used files on Fortress. Your home directory becomes your current working directory, by default, when you log in.
Daily snapshots of your home directory are provided for a limited period of time in the event of accidental deletion. For additional security, you should store another copy of your files on more permanent storage, such as the Fortress HPSS Archive.
Your home directory physically resides on a GPFS storage system in the data center. To find the path to your home directory, first log in then immediately enter the following:
$ pwd
/home/myusername
Or from any subdirectory:
$ echo $HOME
/home/myusername
Your home directory has a quota limiting the total size of files you may store within. For more information, refer to the Storage Quotas / Limits Section.
Link to section 'Lost File Recovery' of 'Home Directory' Lost File Recovery
Nightly snapshots for 7 days, weekly snapshots for 4 weeks, and monthly snapshots for 3 months are kept. This means you will find snapshots from the last 7 nights, the last 4 Sundays, and the last 3 first of the months. Files are available going back between two and three months, depending on how long ago the last first of the month was. Snapshots beyond this are not kept. For additional security, you should store another copy of your files on more permanent storage, such as the Fortress HPSS Archive.
Link to section 'Performance' of 'Home Directory' Performance
Your home directory is medium-performance, non-purged space suitable for tasks like sharing data, editing files, developing and building software, and many other uses.
Your home directory is not designed or intended for use as high-performance working space for running data-intensive jobs with heavy I/O demands.
Scratch Space
Scratch directories are provided for short-term file storage only. The quota of your scratch directory is much greater than the quota of your home directory. You should use your scratch directory for storing temporary input files which your job reads or for writing temporary output files which you may examine after execution of your job. You should use your home directory and Fortress for longer-term storage or for holding critical results. The hsi and htar commands provide easy-to-use interfaces into the archive and can be used to copy files into the archive interactively or even automatically at the end of your regular job submission scripts.
Files in scratch directories are not recoverable. Files in scratch directories are not backed up. If you accidentally delete a file, a disk crashes, or old files are purged, they cannot be restored.
Files are purged from scratch directories not accessed or had content modified in 60 days. Owners of these files receive a notice one week before removal via email. Be sure to regularly check your Purdue email account or set up mail forwarding to an email account you do regularly check. For more information, please refer to our Scratch File Purging Policy.
All users may access scratch directories on Gautschi. To find the path to your scratch directory:
$ findscratch
/scratch/gautschi/myusername
The value of variable $RCAC_SCRATCH is your scratch directory path. Use this variable in any scripts. Your actual scratch directory path may change without warning, but this variable will remain current.
$ echo $RCAC_SCRATCH
/scratch/gautschi/myusername
Scratch directories are specific per cluster. I.e. only the /scratch/gautschi directory is available on Gautschi front-end and compute nodes. No other scratch directories are available on Gautschi.
Your scratch directory has a quota capping the total size and number of files you may store in it. For more information, refer to the section Storage Quotas / Limits.
Link to section 'Performance' of 'Scratch Space' Performance
Your scratch directory is located on a high-performance, large-capacity parallel filesystem engineered to provide work-area storage optimized for a wide variety of job types. It is designed to perform well with data-intensive computations, while scaling well to large numbers of simultaneous connections.
/tmp Directory
/tmp directories are provided for short-term file storage only. Each front-end and compute node has a /tmp directory. Your program may write temporary data to the /tmp directory of the compute node on which it is running. That data is available for as long as your program is active. Once your program terminates, that temporary data is no longer available. When used properly, /tmp may provide faster local storage to an active process than any other storage option. You should use your home directory and Fortress for longer-term storage or for holding critical results.
Backups are not performed for the /tmp directory and removes files from /tmp whenever space is low or whenever the system needs a reboot. In the event of a disk crash or file purge, files in /tmp are not recoverable. You should copy any important files to more permanent storage.
Link to section 'Long-Term Storage' of 'Long-Term Storage' Long-Term Storage
Long-term Storage or Permanent Storage is available to users on the High Performance Storage System (HPSS), an archival storage system, called Fortress. Program files, data files and any other files which are not used often, but which must be saved, can be put in permanent storage. Fortress currently has over 10PB of capacity.
For more information about Fortress, how it works, and user guides, and how to obtain an account:
File Transfer
Gautschi supports several methods for file transfer. Use the links below to learn more about these methods.
SCP
SCP (Secure CoPy) is a simple way of transferring files between two machines that use the SSH protocol. SCP is available as a protocol choice in some graphical file transfer programs and also as a command line program on most Linux, Unix, and Mac OS X systems. SCP can copy single files, but will also recursively copy directory contents if given a directory name.
After Aug 17, 2020, the community clusters will not support password-based authentication for login. Methods that can be used include two-factor authentication (Purdue Login) or SSH keys. If you do not have SSH keys installed, you would need to type your Purdue Login response into the SFTP's "Password" prompt.
Link to section 'Command-line usage:' of 'SCP' Command-line usage:
You can transfer files both to and from Gautschi while initiating an SCP session on either some other computer or on Gautschi (in other words, directionality of connection and directionality of data flow are independent from each other). The scp command appears somewhat similar to the familiar cp command, with an extra user@host:file syntax to denote files and directories on a remote host. Either Gautschi or another computer can be a remote.
-
Example: Initiating SCP session on some other computer (i.e. you are on some other computer, connecting to Gautschi):
(transfer TO Gautschi) (Individual files) $ scp sourcefile myusername@gautschi.rcac.purdue.edu:somedir/destinationfile $ scp sourcefile myusername@gautschi.rcac.purdue.edu:somedir/ (Recursive directory copy) $ scp -pr sourcedirectory/ myusername@gautschi.rcac.purdue.edu:somedir/ (transfer FROM Gautschi) (Individual files) $ scp myusername@gautschi.rcac.purdue.edu:somedir/sourcefile destinationfile $ scp myusername@gautschi.rcac.purdue.edu:somedir/sourcefile somedir/ (Recursive directory copy) $ scp -pr myusername@gautschi.rcac.purdue.edu:sourcedirectory somedir/The -p flag is optional. When used, it will cause the transfer to preserve file attributes and permissions. The -r flag is required for recursive transfers of entire directories.
-
Example: Initiating SCP session on Gautschi (i.e. you are on Gautschi, connecting to some other computer):
(transfer TO Gautschi) (Individual files) $ scp myusername@$another.computer.example.com:sourcefile somedir/destinationfile $ scp myusername@$another.computer.example.com:sourcefile somedir/ (Recursive directory copy) $ scp -pr myusername@$another.computer.example.com:sourcedirectory/ somedir/ (transfer FROM Gautschi) (Individual files) $ scp somedir/sourcefile myusername@$another.computer.example.com:destinationfile $ scp somedir/sourcefile myusername@$another.computer.example.com:somedir/ (Recursive directory copy) $ scp -pr sourcedirectory myusername@$another.computer.example.com:somedir/The -p flag is optional. When used, it will cause the transfer to preserve file attributes and permissions. The -r flag is required for recursive transfers of entire directories.
Link to section 'Software (SCP clients)' of 'SCP' Software (SCP clients)
Linux and other Unix-like systems:
- The
scpcommand-line program should already be installed.
Microsoft Windows:
- MobaXterm
Free, full-featured, graphical Windows SSH, SCP, and SFTP client. - Command-line
scpprogram can be installed as part of Windows Subsystem for Linux (WSL), or Git-Bash.
Mac OS X:
- The
scpcommand-line program should already be installed. You may start a local terminal window from "Applications->Utilities". - Cyberduck is a full-featured and free graphical SFTP and SCP client.
FTP / SFTP
FTP is not supported on any research systems because it does not allow for secure transmission of data. Use SFTP instead, as described below.
SFTP (Secure File Transfer Protocol) is a reliable way of transferring files between two machines. SFTP is available as a protocol choice in some graphical file transfer programs and also as a command-line program on most Linux, Unix, and Mac OS X systems. SFTP has more features than SCP and allows for other operations on remote files, remote directory listing, and resuming interrupted transfers. Command-line SFTP cannot recursively copy directory contents; to do so, try using SCP or graphical SFTP client.
After Aug 17, 2020, the community clusters will not support password-based authentication for login. Methods that can be used include two-factor authentication (Purdue Login) or SSH keys. If you do not have SSH keys installed, you would need to type your Purdue Login response into the SFTP's "Password" prompt.
Link to section 'Command-line usage' of 'FTP / SFTP' Command-line usage
You can transfer files both to and from Gautschi while initiating an SFTP session on either some other computer or on Gautschi (in other words, directionality of connection and directionality of data flow are independent from each other). Once the connection is established, you use put or get subcommands between "local" and "remote" computers. Either Gautschi or another computer can be a remote.
-
Example: Initiating SFTP session on some other computer (i.e. you are on another computer, connecting to Gautschi):
$ sftp myusername@gautschi.rcac.purdue.edu (transfer TO Gautschi) sftp> put sourcefile somedir/destinationfile sftp> put -P sourcefile somedir/ (transfer FROM Gautschi) sftp> get sourcefile somedir/destinationfile sftp> get -P sourcefile somedir/ sftp> exitThe -P flag is optional. When used, it will cause the transfer to preserve file attributes and permissions.
-
Example: Initiating SFTP session on Gautschi (i.e. you are on Gautschi, connecting to some other computer):
$ sftp myusername@$another.computer.example.com (transfer TO Gautschi) sftp> get sourcefile somedir/destinationfile sftp> get -P sourcefile somedir/ (transfer FROM Gautschi) sftp> put sourcefile somedir/destinationfile sftp> put -P sourcefile somedir/ sftp> exitThe -P flag is optional. When used, it will cause the transfer to preserve file attributes and permissions.
Link to section 'Software (SFTP clients)' of 'FTP / SFTP' Software (SFTP clients)
Linux and other Unix-like systems:
- The
sftpcommand-line program should already be installed.
Microsoft Windows:
- MobaXterm
Free, full-featured, graphical Windows SSH, SCP, and SFTP client. - Command-line
sftpprogram can be installed as part of Windows Subsystem for Linux (WSL), or Git-Bash.
Mac OS X:
- The
sftpcommand-line program should already be installed. You may start a local terminal window from "Applications->Utilities". - Cyberduck is a full-featured and free graphical SFTP and SCP client.
Globus
Link to section 'Globus' of 'Globus' Globus
Globus, previously known as Globus Online, is a powerful and easy to use file transfer service for transferring files virtually anywhere. It works within RCAC's various research storage systems; it connects between RCAC and remote research sites running Globus; and it connects research systems to personal systems. You may use Globus to connect to your home, scratch, and Fortress storage directories. Since Globus is web-based, it works on any operating system that is connected to the internet. The Globus Personal client is available on Windows, Linux, and Mac OS X. It is primarily used as a graphical means of transfer but it can also be used over the command line.
Link to section 'Link to section 'Globus Web:' of 'Globus' Globus Web:' of 'Globus' Link to section 'Globus Web:' of 'Globus' Globus Web:
- Navigate to http://transfer.rcac.purdue.edu
- Click "Proceed" to log in with your Purdue Career Account.
- On your first login it will ask to make a connection to a Globus account. Accept the conditions.
- Now you are at the main screen. Click "File Transfer" which will bring you to a two-panel interface (if you only see one panel, you can use selector in the top-right corner to switch the view).
- You will need to select one collection and file path on one side as the source, and the second collection on the other as the destination. This can be one of several Purdue endpoints, or another University, or even your personal computer (see Personal Client section below).
The RCAC collections are as follows. A search for "Purdue" will give you several suggested results you can choose from, or you can give a more specific search.
- Home directory and Scratch storage: "Gautschi Cluster Collection", however, you can start typing "gautschi" and it will suggest appropriate matches.
- Research Data Depot: "Purdue Research Computing - Data Depot", a search for "Depot" should provide appropriate matches to choose from.
- Fortress: "Purdue Fortress HPSS Archive", a search for "Fortress" should provide appropriate matches to choose from.
From here, select a file or folder in either side of the two-pane window, and then use the arrows in the top-middle of the interface to instruct Globus to move files from one side to the other. You can transfer files in either direction. You will receive an email once the transfer is completed.
Link to section 'Link to section 'Globus Personal Client setup:' of 'Globus' Globus Personal Client setup:' of 'Globus' Link to section 'Globus Personal Client setup:' of 'Globus' Globus Personal Client setup:
Globus Connect Personal is a small software tool you can install to make your own computer a Globus endpoint on its own. It is useful if you need to transfer files via Globus to and from your computer directly.
- On the "Collections" page from earlier, click "Get Globus Connect Personal" or download a version for your operating system it from here: Globus Connect Personal
- Name this particular personal system and follow the setup prompts to create your Globus Connect Personal endpoint.
- Your personal system is now available as a collection within the Globus transfer interface.
Link to section 'Link to section 'Globus Command Line:' of 'Globus' Globus Command Line:' of 'Globus' Link to section 'Globus Command Line:' of 'Globus' Globus Command Line:
Globus supports command line interface, allowing advanced automation of your transfers.
To use the recommended standalone Globus CLI application (the globus command):
- First time use: issue the globus login command and follow instructions for initial login.
- Commands for interfacing with the CLI can be found via Using the Command Line Interface, as well as the Globus CLI Examples pages.
Link to section 'Link to section 'Sharing Data with Outside Collaborators' of 'Globus' Sharing Data with Outside Collaborators' of 'Globus' Link to section 'Sharing Data with Outside Collaborators' of 'Globus' Sharing Data with Outside Collaborators
Globus allows convenient sharing of data with outside collaborators. Data can be shared with collaborators' personal computers or directly with many other computing resources at other institutions. See the Globus documentation on how to share data:
For links to more information, please see Globus Support page and RCAC Globus presentation.
Windows Network Drive / SMB
SMB (Server Message Block), also known as CIFS, is an easy-to-use file transfer protocol that is useful for transferring files between RCAC systems and a desktop or laptop. You may use SMB to connect to your home, scratch, and Fortress storage directories. The SMB protocol is available on Windows, Linux, and Mac OS X. It is primarily used as a graphical means of transfer, but it can also be used over the command line.
Note: to access Gautschi through SMB file sharing, you must be on a Purdue campus network or connected through VPN.
Link to section 'Windows:' of 'Windows Network Drive / SMB' Windows:
- Windows 7: Click Windows menu > Computer, then click Map Network Drive in the top bar
- Windows 8 & 10: Tap the Windows key, type computer, select This PC, click Computer > Map Network Drive in the top bar
- Windows 11: Tap the Windows key, type File Explorer, select This PC, click Computer > Map Network Drive in the top bar
- In the folder location, enter the following information and click Finish:
- To access your Gautschi home directory, enter \\home.gautschi.rcac.purdue.edu\gautschi-home.
- To access your scratch space on Gautschi, enter \\scratch.gautschi.rcac.purdue.edu\gautschi-scratch. Once mapped, you will be able to navigate to your scratch directory.
- Note: Use your career account login name and password when prompted. (You will not need to add "
,push" nor use your Purdue Duo client.) - Your home or scratch directory should now be mounted as a drive in the Computer window.
- If you have issues mapping the drive check Credential Manager.
- In the Search bar, type "Credential Manager"
- Inside Credential Manager, click "Windows Credentials" and "Add New Credential"
- Servername:
\\home.negishi.rcac.purdue.edu - Username:
boilerad\PurdueLogin - Then save it and try mapping again.
-
If there's already an entry for the server you are trying to map delete it.
-
Link to section 'Mac OS X:' of 'Windows Network Drive / SMB' Mac OS X:
- In the Finder, click Go > Connect to Server
- In the Server Address enter the following information and click Connect:
- To access your Gautschi home directory, enter smb://home.gautschi.rcac.purdue.edu/gautschi-home.
- To access your scratch space on Gautschi, enter smb://scratch.gautschi.rcac.purdue.edu/gautschi-scratch. Once mapped, you will be able to navigate to your scratch directory.
- Note: Use your career account login name and password when prompted. (You will not need to add "
,push" nor use your Purdue Duo client.) - Your home or scratch directory should now be mounted as a drive in the Computer window.
Link to section 'Linux:' of 'Windows Network Drive / SMB' Linux:
- There are several graphical methods to connect in Linux depending on your desktop environment. Once you find out how to connect to a network server on your desktop environment, choose the Samba/SMB protocol and adapt the information from the Mac OS X section to connect.
- If you would like access via samba on the command line you may install smbclient which will give you FTP-like access and can be used as shown below. For all the possible ways to connect look at the Mac OS X instructions.
smbclient //home.gautschi.rcac.purdue.edu/gautschi-home -U boilerad/myusername smbclient //scratch.gautschi.rcac.purdue.edu/gautschi-scratch -U boilerad/myusername - Note: Use your career account login name and password when prompted. (You will not need to add "
,push" nor use your Purdue Duo client.)
HSI
HSI, the Hierarchical Storage Interface, is the preferred method of transferring files to and from Gautschi. HSI is designed to be a friendly interface for users of the High Performance Storage System (HPSS). It provides a familiar Unix-style environment for working within HPSS while automatically taking advantage of high-speed, parallel file transfers without requiring any special user knowledge.
HSI is provided on all research systems as the command hsi. HSI is also available for download for many operating systems.
Interactive usage:
$ hsi
*************************************************************************
* Purdue University
* High Performance Storage System (HPSS)
*************************************************************************
* This is the Purdue Data Archive, Fortress. For further information
* see http://www.rcac.purdue.edu/storage/fortress/
*
* If you are having problems with HPSS, please call IT/Operational
* Services at 49-44000 or send E-mail to rcac-help@purdue.edu.
*
*************************************************************************
Username: myusername UID: 12345 Acct: 12345(12345) Copies: 1 Firewall: off [hsi.3.5.8 Wed Sep 21 17:31:14 EDT 2011]
[Fortress HSI]/home/myusername->put data1.fits
put 'test' : '/home/myusername/test' ( 1024000000 bytes, 250138.1 KBS (cos=11))
[Fortress HSI]/home/myusername->lcd /tmp
[Fortress HSI]/home/myusername->get data1.fits
get '/tmp/data1.fits' : '/home/myusername/data1.fits' (2011/10/04 16:28:50 1024000000 bytes, 325844.9 KBS )
[Fortress HSI]/home/myusername->quit
Batch transfer file:
put data1.fits
put data2.fits
put data3.fits
put data4.fits
put data5.fits
put data6.fits
put data7.fits
put data8.fits
put data9.fits
Batch usage:
$ hsi < my_batch_transfer_file
*************************************************************************
* Purdue University
* High Performance Storage System (HPSS)
*************************************************************************
* This is the Purdue Data Archive, Fortress. For further information
* see http://www.rcac.purdue.edu/storage/fortress/
*
* If you are having problems with HPSS, please call IT/Operational
* Services at 49-44000 or send E-mail to rcac-help@purdue.edu.
*
*************************************************************************
Username: myusername UID: 12345 Acct: 12345(12345) Copies: 1 Firewall: off [hsi.3.5.8 Wed Sep 21 17:31:14 EDT 2011]
put 'data1.fits' : '/home/myusername/data1.fits' ( 1024000000 bytes, 250200.7 KBS (cos=11))
put 'data2.fits' : '/home/myusername/data2.fits' ( 1024000000 bytes, 258893.4 KBS (cos=11))
put 'data3.fits' : '/home/myusername/data3.fits' ( 1024000000 bytes, 222819.7 KBS (cos=11))
put 'data4.fits' : '/home/myusername/data4.fits' ( 1024000000 bytes, 224311.9 KBS (cos=11))
put 'data5.fits' : '/home/myusername/data5.fits' ( 1024000000 bytes, 323707.3 KBS (cos=11))
put 'data6.fits' : '/home/myusername/data6.fits' ( 1024000000 bytes, 320322.9 KBS (cos=11))
put 'data7.fits' : '/home/myusername/data7.fits' ( 1024000000 bytes, 253192.6 KBS (cos=11))
put 'data8.fits' : '/home/myusername/data8.fits' ( 1024000000 bytes, 253056.2 KBS (cos=11))
put 'data9.fits' : '/home/myusername/data9.fits' ( 1024000000 bytes, 323218.9 KBS (cos=11))
EOF detected on TTY - ending HSI session
For more information about HSI:
HTAR
HTAR (short for "HPSS TAR") is a utility program that writes TAR-compatible archive files directly onto Gautschi, without having to first create a local file. Its command line was originally based on tar, with a number of extensions added to provide extra features.
HTAR is provided on all research systems as the command htar. HTAR is also available for download for many operating systems.
Link to section 'Usage:' of 'HTAR' Usage:
Create a tar archive on Gautschi named data.tar including all files with the extension ".fits":
$ htar -cvf data.tar *.fits
HTAR: a data1.fits
HTAR: a data2.fits
HTAR: a data3.fits
HTAR: a data4.fits
HTAR: a data5.fits
HTAR: a /tmp/HTAR_CF_CHK_17953_1317760775
HTAR Create complete for data.tar. 5,120,006,144 bytes written for 5 member files, max threads: 3 Transfer time: 16.457 seconds (311.121 MB/s)
HTAR: HTAR SUCCESSFUL
Unpack a tar archive on Gautschi named data.tar into a scratch directory for use in a batch job:
$ cd $RCAC_SCRATCH/job_dir
$ htar -xvf data.tar
HTAR: x data1.fits, 1024000000 bytes, 2000001 media blocks
HTAR: x data2.fits, 1024000000 bytes, 2000001 media blocks
HTAR: x data3.fits, 1024000000 bytes, 2000001 media blocks
HTAR: x data4.fits, 1024000000 bytes, 2000001 media blocks
HTAR: x data5.fits, 1024000000 bytes, 2000001 media blocks
HTAR: Extract complete for data.tar, 5 files. total bytes read: 5,120,004,608 in 18.841 seconds (271.749 MB/s )
HTAR: HTAR SUCCESSFUL
Look at the contents of the data.tar HTAR archive on Gautschi:
$ htar -tvf data.tar
HTAR: -rw-r--r-- myusername/pucc 1024000000 2011-10-04 16:30 data1.fits
HTAR: -rw-r--r-- myusername/pucc 1024000000 2011-10-04 16:35 data2.fits
HTAR: -rw-r--r-- myusername/pucc 1024000000 2011-10-04 16:35 data3.fits
HTAR: -rw-r--r-- myusername/pucc 1024000000 2011-10-04 16:35 data4.fits
HTAR: -rw-r--r-- myusername/pucc 1024000000 2011-10-04 16:35 data5.fits
HTAR: -rw------- myusername/pucc 256 2011-10-04 16:39 /tmp/HTAR_CF_CHK_17953_1317760775
HTAR: Listing complete for data.tar, 6 files 6 total objects
HTAR: HTAR SUCCESSFUL
Unpack a single file, "data5.fits", from the tar archive on Gautschi named data.tar into a scratch directory.:
$ htar -xvf data.tar data5.fits
HTAR: x data5.fits, 1024000000 bytes, 2000001 media blocks
HTAR: Extract complete for data.tar, 1 files. total bytes read: 1,024,000,512 in 3.642 seconds (281.166 MB/s )
HTAR: HTAR SUCCESSFUL
Link to section 'HTAR Archive Verification' of 'HTAR' HTAR Archive Verification
HTAR allows different types of content verification while creating archives. Users can ask HTAR to verify the contents of an archive during (or after) creation using the '-Hverify' switch. The syntax of this option is:
$ htar -Hverify=option[,option...] ... other arguments ...
| Option | Explanation |
|---|---|
info |
Compares tar header info with the corresponding values in the index. |
crc |
Enables CRC checking of archive files for which a CRC was generated when the file is added to the archive. |
compare |
Enables a byte-by-byte comparison of archive member files and their local file counterparts. |
nocrc |
Disables CRC checking of archive files. |
nocompare |
Disables a byte-by-byte comparison of archive member files and their local file counterparts. |
Users can use a comma-separated list of options shown above, or a numeric value, or the wildcard all to specify the degree of verification. The numeric values for Hverify can be interpreted as follows:
0: Enables "info" verification.
1: Enables level 0 + "crc" verification.
2: Enables level 1 + "compare" verification.
all: Enables all comparison options.
An example to verify an archive during creation using checksums (crc):
htar -Hverify=1 -cvf abc.tar ./abc
An example to verify a previously created archive using checksums (crc):
htar -Hverify=1 -Kvf abc.tar
Please note that the time for verifying an archive increases as you increase the verification level. Carefully choose the option that suits your dataset best.
For details please see the HTAR Man Page.
For more information about HTAR:
HTAR has an individual file size limit of 64GB. If any files you are trying to archive with HTAR are greater than 64GB, then HTAR will immediately fail. This does not limit the number of files in the archive or the total overall size of the archive. To get around this limitation, try using the htar_large command. It is slower than using htar but it will work around the 64GB file size limit. This does not limit the number of files in the archive or the total overall size of the archive.
To get around this limitation, try using the htar_large command. It is slower than using HTAR but it will work around the 64GB file size limit. The usage of htar_large is almost the same as htar except that htar_large will not generate the tar index file. Thus, the -Hverify=1 option cannot be used since it's based on index file.
Storage Quota / Limits
Some limits are imposed on your disk usage on research systems. A quota is implemented on each filesystem. Each filesystem (home directory, scratch directory, etc.) may have a different limit. If you exceed the quota, you will not be able to save new files or new data to the filesystem until you delete or move data to long-term storage.
Link to section 'Checking Quota' of 'Storage Quota / Limits' Checking Quota
To check the current quotas of your home and scratch directories check the My Quota page or use the myquota command:
$ myquota
Type Filesystem Size Limit Use Files Limit Use
==============================================================================
home myusername 5.0GB 25.0GB 20% - - -
scratch gautschi 220.7GB 100.0TB 0.22% 8k 2,000k 0.43%
The columns are as follows:
- Type: indicates home or scratch directory or your depot space.
- Filesystem: name of storage option.
- Size: sum of file sizes in bytes.
- Limit: allowed maximum on sum of file sizes in bytes.
- Use: percentage of file-size limit currently in use.
- Files: number of files and directories (not the size).
- Limit: allowed maximum on number of files and directories. It is possible, though unlikely, to reach this limit and not the file-size limit if you create a large number of very small files.
- Use: percentage of file-number limit currently in use.
If you find that you reached your quota in either your home directory or your scratch file directory, obtain estimates of your disk usage. Find the top-level directories which have a high disk usage, then study the subdirectories to discover where the heaviest usage lies.
To see in a human-readable format an estimate of the disk usage of your top-level directories in your home directory:
$ du -h --max-depth=1 $HOME >myfile
32K /home/myusername/mysubdirectory_1
529M /home/myusername/mysubdirectory_2
608K /home/myusername/mysubdirectory_3
The second directory is the largest of the three, so apply command du to it.
To see in a human-readable format an estimate of the disk usage of your top-level directories in your scratch file directory:
$ du -h --max-depth=1 $RCAC_SCRATCH >myfile
160K /scratch/gautschi/myusername
This strategy can be very helpful in figuring out the location of your largest usage. Move unneeded files and directories to long-term storage to free space in your home and scratch directories.
Link to section 'Increasing Quota' of 'Storage Quota / Limits' Increasing Quota
Link to section 'Home Directory' of 'Storage Quota / Limits' Home Directory
If you find you need additional disk space in your home directory, please consider archiving and compressing old files and moving them to long-term storage on the Fortress HPSS Archive, or purchase the Depot space for long-term storage. Unfortunately, it is not possible to increase your home directory quota beyond it's current level.
Link to section 'Scratch Space' of 'Storage Quota / Limits' Scratch Space
If you find you need additional disk space in your scratch space, please first consider archiving and compressing old files and moving them to long-term storage on the Fortress HPSS Archive. If you are unable to do so, you may ask for a quota increase by contacting support.
Link to section 'Storage Environment Variables' of 'Storage Environment Variables' Storage Environment Variables
Several environment variables are automatically defined for you to help you manage your storage. Use environment variables instead of actual paths whenever possible to avoid problems if the specific paths to any of these change.
| Name | Description |
|---|---|
| HOME | /home/myusername |
| PWD | path to your current directory |
| RCAC_SCRATCH | /scratch/gautschi/myusername |
By convention, environment variable names are all uppercase. You may use them on the command line or in any scripts in place of and in combination with hard-coded values:
$ ls $HOME
...
$ ls $RCAC_SCRATCH/myproject
...
To find the value of any environment variable:
$ echo $RCAC_SCRATCH
/scratch/gautschi/myusername
To list the values of all environment variables:
$ env
USER=myusername
HOME=/home/myusername
RCAC_SCRATCH=/scratch/gautschi/myusername
...
You may create or overwrite an environment variable. To pass (export) the value of a variable in bash:
$ export MYPROJECT=$RCAC_SCRATCH/myproject
To assign a value to an environment variable in either tcsh or csh:
$ setenv MYPROJECT value
Link to section 'Archive and Compression' of 'Archive and Compression' Archive and Compression
There are several options for archiving and compressing groups of files or directories. The mostly commonly used options are:
Link to section 'tar' of 'Archive and Compression' tar
See the official documentation for tar for more information.
Saves many files together into a single archive file, and restores individual files from the archive. Includes automatic archive compression/decompression options and special features for incremental and full backups.
Examples:
(list contents of archive somefile.tar)
$ tar tvf somefile.tar
(extract contents of somefile.tar)
$ tar xvf somefile.tar
(extract contents of gzipped archive somefile.tar.gz)
$ tar xzvf somefile.tar.gz
(extract contents of bzip2 archive somefile.tar.bz2)
$ tar xjvf somefile.tar.bz2
(archive all ".c" files in current directory into one archive file)
$ tar cvf somefile.tar *.c
(archive and gzip-compress all files in a directory into one archive file)
$ tar czvf somefile.tar.gz somedirectory/
(archive and bzip2-compress all files in a directory into one archive file)
$ tar cjvf somefile.tar.bz2 somedirectory/
Other arguments for tar can be explored by using the man tar command.
Link to section 'gzip' of 'Archive and Compression' gzip
The standard compression system for all GNU software.
Examples:
(compress file somefile - also removes uncompressed file)
$ gzip somefile
(uncompress file somefile.gz - also removes compressed file)
$ gunzip somefile.gz
Link to section 'bzip2' of 'Archive and Compression' bzip2
See the official documentation for bzip for more information.
Strong, lossless data compressor based on the Burrows-Wheeler transform. Stronger compression than gzip.
Examples:
(compress file somefile - also removes uncompressed file)
$ bzip2 somefile
(uncompress file somefile.bz2 - also removes compressed file)
$ bunzip2 somefile.bz2
There are several other, less commonly used, options available as well:
- zip
- 7zip
- xz
Link to section 'Sharing Files from Gautschi' of 'Sharing' Sharing Files from Gautschi
Gautschi supports several methods for file sharing. Use the links below to learn more about these methods.
Link to section 'Sharing Data with Globus' of 'Globus' Sharing Data with Globus
Data on any RCAC resource can be shared with other users within Purdue or with collaborators at other institutions. Globus allows convenient sharing of data with outside collaborators. Data can be shared with collaborators' personal computers or directly with many other computing resources at other institutions.
To share files, login to https://transfer.rcac.purdue.edu, navigate to the endpoint (collection) of your choice, and follow instructions as described in Globus documentation on how to share data:
See also RCAC Globus presentation.
Link to section 'Sharing static content from your Data Depot space via the WWW' of 'WWW' Sharing static content from your Data Depot space via the WWW
Your research group can easily share static files (images, data, HTML) from your depot space via the WWW.
- Contact support to set up a "www" folder in your Data Depot space.
- Copy any files that you wish to share via the WWW into your Data Depot space's "www" folder.
- For example, cp /path/to/image.jpg /depot/mylab/www/, where mylab is your research group name.
- or upload to smb://datadepot.rcac.purdue.edu/depot/mylab/www, where mylab is your research group name.
- Your file is now accessible via your web browser at the URL https://www.datadepot.rcac.purdue.edu/mylab/image.jpg
Note that it is not possible to run web sites, dynamic content, interpreters (PHP, Perl, Python), or CGI scripts from this web site.
Gateway (Open OnDemand)
Gautschi's Gateway is an open-source HPC portal developed by the Ohio Supercomputing Center. Open OnDemand allows one to interact with HPC resources through a web browser and easily manage files, submit jobs, and interact with graphical applications directly in a browser, all with no software to install. Gautschi has an instance of OnDemand available that can be accessed via gateway.gautschi.rcac.purdue.edu.
Link to section 'Logging In' of 'Gateway (Open OnDemand)' Logging In
To log into Gateway:
- Navigate to gateway.gautschi.rcac.purdue.edu
- Log in using your Career account username and Purdue Login Duo client.
On the splash page you will see a quota usage report. If you are over 90% on any of your quotas a warning will be displayed. This information will update every 10-15 minutes while you are active on Gateway.
Link to section 'Apps' of 'Gateway (Open OnDemand)' Apps
There are a number of built-in apps in Gateway that can be accessed from the top menu bar. Below are links to documentation on each app.
Files
The Files app will let you access your files in your Home Directory, Scratch, and Data Depot spaces. The app lets you manage create, manage, and delete files and directories from your web browser. Navigate by double clicking on folders in the file explorer or by using the file tree on the left.
On the top row, there are buttons to:
- Go To: directly input a directory to navigate to
- Open in Terminal: launches the Shell app and navigates you to the current directory in the terminal
- New File: creates a new, empty file
- New Dir: creates a new, empty directory
- Upload: upload a file from your computer
Note: File uploads from your browser are limited to 100 GB per file. Be mindful that uploads over a few gigabytes may be unreliable through your browser, especially from off-campus connections. For very large files or off-campus transfers alternative methods such as Globus are highly recommended.
The second row of buttons lets you perform typical file management operations. The Edit button will open files in a fully fledged browser based text editor - it features syntax highlighting and vim and Emacs key bindings.
Jobs
There are two apps under the Jobs apps: Active Jobs and Job Composer. These are detailed below.
Link to section 'Active Jobs' of 'Jobs' Active Jobs
This shows you active SLURM jobs currently on the cluster. The default view will show you your current jobs, similar to squeue -u rices. Using the button labeled "Your Jobs" in the upper right allows you to select different filters by queue (account). All accounts output by slist will appear for you here. Using the arrow on the left hand side will expand the full job details.
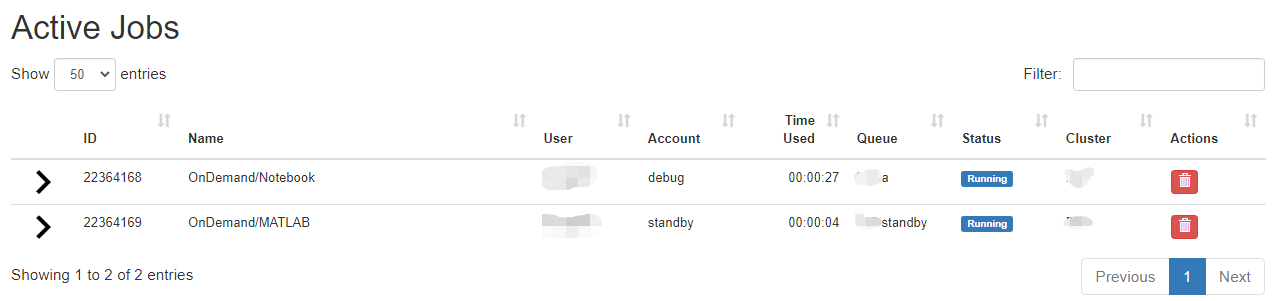
Link to section 'Job Composer' of 'Jobs' Job Composer
The Job Composer app allows you to create and submit jobs to the cluster. You can select from pre-defined templates (most of these are taken from the User Guide examples) or you can create your own templates for frequently used workflows.
Link to section 'Creating Job from Existing Template' of 'Jobs' Creating Job from Existing Template
Click "New Job" menu, then select "From Template":
Then select from one of the available templates.
Click 'Create New Job' in second pane.
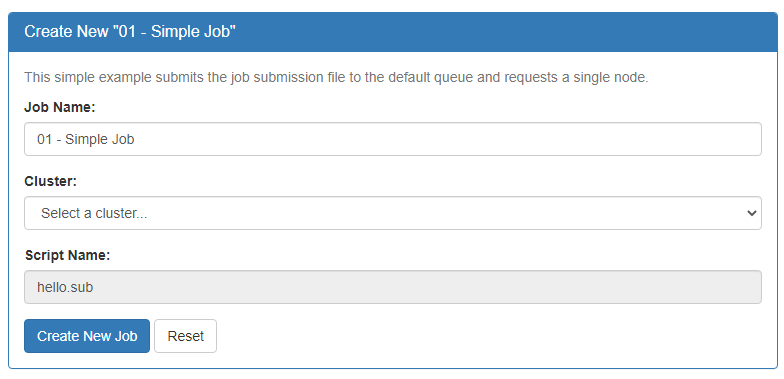
Your new job should be selected in your list of jobs. In the 'Submit Script' pane you can see the job script that was generated with an 'Open Editor' link to open the script in the built-in editor. Open the file in the editor and edit the script as necessary. By default the job will specify standby queue - this should be changed as appropriate, along with the node and walltime requests.
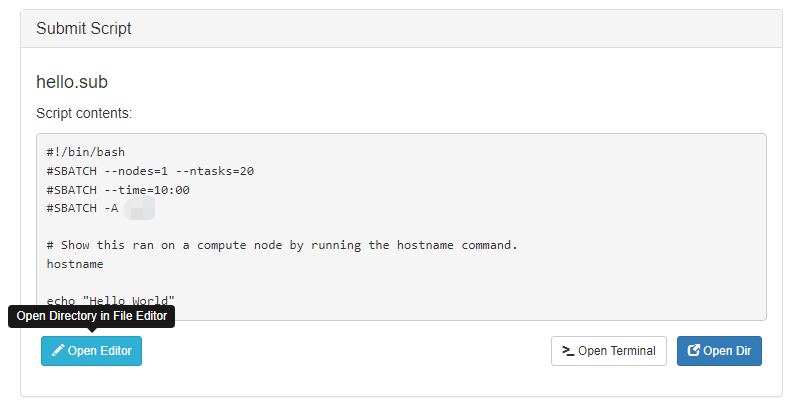
When you are finished with editing the job and are ready to submit, click the green 'Submit' button at the top of the job list. You can monitor progress from here or from the Active Jobs app. Once completed, you should see the output files appear:

Clicking on one of the output files will open it in the file editor for your viewing.
Link to section 'Creating New Template' of 'Jobs' Creating New Template
First, prepare a template directory containing a template submission script along with any input files. Then, to import the job into the Job Composer app, click the 'Create New Template' button. Fill in the directory containing your template job script and files in the first box. Give it an appropriate name and notes.
This template will now appear in your list of templates to choose from when composing jobs. You can now go create and submit a job from this new template.
Cluster Tools
The Cluster Tools menu contains cluster utilities. At the moment, only a terminal app is provided. Additional apps may be developed and provided in the future.
Link to section 'Shell Access' of 'Cluster Tools' Shell Access
Launching the shell app will provide you with a web-based terminal session on the cluster front-end. This is equivalent to using a standalone SSH client to connect to gautschi.rcac.purdue.edu where you are connected to one several front-ends. The normal acceptable front-end use policy applies to access through the web-app. X11 Forwarding is not supported. Use of one of the interactive apps is recommended for graphical applications.
Interactive Apps
There are several interactive apps available through Gateway that can be accessed through the Interactive Apps dropdown menu. These apps are provided with a basic node and software configuration as a 'quick-launch' option to get your work up and running quickly. For simplicity, minimal options are provided - these apps are not intended for complex configuration/customization scenarios.
After you a submit an interactive app to the queue, Gateway will track and manage the session. Once it starts, you may connect and disconnect from the session in your browser, leaving the job running while you log out of your browser.
Each of the available apps are documented through the following links.
Compute Node Desktop
The Compute Node Desktop app will launch a graphical desktop session on a compute node. This is similar to using Thinlinc, however, this gives you a desktop directly on a compute node instead on a front-end. This app is useful if you have a custom application or application not directly available as an interactive app you would like to run inside Gateway.
To launch a desktop session on a compute node, select the Gautschi Compute Desktop app. From the submit form, select from the available options - the queue to which you wish to submit and the number of wallclock hours you wish to have job running. There is also a checkbox that enable a notification to your email when the job starts.
After the interactive job is submitted you will be taken to your list of active interactive app sessions. You can monitor the status of the job from here until it starts, or if you enabled the email notification, watch your Purdue email for the notification the job has started.
Once it is indicated the job has started you can connect to the desktop with the "Launch noVNC in New Tab" button. The session will be terminated after the wallclock hours you specified have elapsed or you terminate the session early with the "Delete" button from the list of sessions. Deleting the session when you are finished will free up queue resources for your lab mates and other users on the system.
MATLAB
The MATLAB app will launch a MATLAB session on a compute node and allow you to connect directly to it in a web browser.
To launch a MATLAB session on a compute node, select the MATLAB app. From the submit form, select from the available options - the version of MATLAB you are interested in running, the queue to which you wish to submit, and the number of wallclock hours you wish to have job running. There is also a checkbox that enable a notification to your email when the job starts.
After the interactive job is submitted you will be taken to your list of active interactive app sessions. You can monitor the status of the job from here until it starts, or if you enabled the email notification, watch your Purdue email for the notification the job has started.
Once it is indicated the job has started you can connect to the desktop with the "Launch noVNC in New Tab" button. The session will be terminated after the wallclock hours you specified have elapsed or you terminate the session early with the "Delete" button from the list of sessions. Deleting the session when you are finished will free up queue resources for your lab mates and other users on the system.
Jupyter Notebook
The Notebook app will launch a Notebook session on a compute node and allow you to connect directly to it in a web browser.
To launch a Notebook session on a compute node, select the Notebook app. From the submit form, select from the available options:
- Queue: This is a dropdown menu from which you can select a queue from all of the queues to which you have permission to submit.
- Walltime: This is a field which expects a number and represents how many hours you want to keep the session running. Note that this value should not exceed the maximum value given next to the selected queue name from the queue dropdown menu.
- Number of Cores/GPUs: This is a field which expects a number and represents the number of your resources your session is requesting. Note that the amount of memory allocated for your session is proportional to the number of cores or GPUs that you request for your job, so if your session is running out of memory, consider increasing this value.
- Use Jupyter Lab: This is a checkbox which, when checked, will run Jupyter Lab instead of Jupyter Notebook. Both of these applications are interfaces to Jupyter, and you can launch Jupyter notebooks from within Jupyter Lab. Jupyter Notebook is more "barebones" while Jupyter Lab has additional features such as the ability to interact with additional file types.
- E-mail Notice: This is a checkbox which, when checked, will send you an e-mail notification to your Purdue e-mail that your session is ready when the scheduler has found resources to dedicate to your session.
After the interactive job is submitted you will be taken to your list of active interactive app sessions. You can monitor the status of the job from here until it starts, or if you enabled the email notification, watch your Purdue email for the notification the job has started.
Once it is indicated the job has started you can connect to the desktop with the "Connect to Jupyter" button. Once connected, you can create new notebooks, selecting the currently available Anaconda versions available as modules, and any personally created Notebook kernels.
Often times you may want to use one of your existing Anaconda environments within your Jupyter session to use libraries specific to your workflow. In order to do so, you must ensure that the Anaconda environment you want to use contains the Python packages "IPyKernel" and "IPython" which are packages that are required by Jupyter. When you create a Jupyter session, Open OnDemand will check through your existing Anaconda environments and create a Jupyter kernel for any Anaconda environment that contains these two packages, and you will be able to select to use that kernel from within the application.
The session will be terminated after the number of hours you specified have elapsed or you terminate the session early with the "Delete" button from the list of sessions. Deleting the session when you are finished will free up queue resources for your lab mates and other users on the system.
RStudio Server
The RStudio app will launch a RStudio session on a compute node and allow you to connect directly to it in a web browser.
To launch a RStudio session on a compute node, select the RStudio app. From the submit form, select from the available options - the queue to which you wish to submit, and the number of wallclock hours you wish to have job running. There is also a checkbox that enable a notification to your email when the job starts.
After the interactive job is submitted you will be taken to your list of active interactive app sessions. You can monitor the status of the job from here until it starts, or if you enabled the email notification, watch your Purdue email for the notification the job has started.
Once it is indicated the job has started you can connect to the desktop with the "Connect to RStudio Server" button. The session will be terminated after the wallclock hours you specified have elapsed or you terminate the session early with the "Delete" button from the list of sessions. Deleting the session when you are finished will free up queue resources for your lab mates and other users on the system.
Compiling Source Code
Documentation on compiling source code on Gautschi.
Compiling GPU Programs
The Gautschi cluster nodes contain 6 GPUs that support CUDA and OpenCL. See the detailed hardware overview for the specifics on the GPUs in Gautschi. This section focuses on using CUDA.
A simple CUDA program has a basic workflow:
- Initialize an array on the host (CPU).
- Copy array from host memory to GPU memory.
- Apply an operation to array on GPU.
- Copy array from GPU memory to host memory.
Here is a sample CUDA program:
Both front-ends and GPU-enabled compute nodes have the CUDA tools and libraries available to compile CUDA programs. To compile a CUDA program, load CUDA, and use nvcc to compile the program:
$ module load gcc/11.4.1 cuda/12.6.0
$ nvcc gpu_hello.cu -o gpu_hello
./gpu_hello
No GPU specified, using first GPUhello, world
The example illustrates only how to copy an array between a CPU and its GPU but does not perform a serious computation.
The following program times three square matrix multiplications on a CPU and on the global and shared memory of a GPU:
$ module load cuda
$ nvcc mm.cu -o mm
$ ./mm 0
speedup
-------
Elapsed time in CPU: 6555.2 milliseconds
Elapsed time in GPU (global memory): 32.9 milliseconds 199.1
Elapsed time in GPU (shared memory): 3.0 milliseconds 2191.8
For best performance, the input array or matrix must be sufficiently large to overcome the overhead in copying the input and output data to and from the GPU.
For more information about NVIDIA, CUDA, and GPUs:
Compiling Hybrid Programs
A hybrid program combines both MPI and shared-memory to take advantage of compute clusters with multi-core compute nodes. Libraries for OpenMPI and Intel MPI (IMPI) and compilers which include OpenMP for C, C++, and Fortran are available.
| Language | Header Files |
|---|---|
| Fortran 77 |
|
| Fortran 90 |
|
| Fortran 95 |
|
| C |
|
| C++ |
|
A few examples illustrate hybrid programs with task parallelism of OpenMP:
This example illustrates a hybrid program with loop-level (data) parallelism of OpenMP:
To see the available MPI libraries:
$ module avail impi
$ module avail openmpi
The following tables illustrate how to compile your hybrid (MPI/OpenMP) program. Any compiler flags accepted by Intel ifort/icc compilers are compatible with their respective MPI compiler.
| Language | Command |
|---|---|
| Fortran 77 |
|
| Fortran 90 |
|
| Fortran 95 |
|
| C |
|
| C++ |
|
| Language | Command |
|---|---|
| Fortran 77 |
|
| Fortran 90 |
|
| Fortran 95 |
|
| C |
|
| C++ |
|
The Intel and GNU compilers will not output anything for a successful compilation. Also, the Intel compiler does not recognize the suffix .f95.
Compiling Serial Programs
A serial program is a single process which executes as a sequential stream of instructions on one processor core. Compilers capable of serial programming are available for C, C++, and versions of Fortran.
Here are a few sample serial programs:
- serial_hello.f
- serial_hello.f90
- serial_hello.f95
- serial_hello.c
-
To load a compiler, enter one of the following:
$ module load intel
$ module load gcc
| Language | Intel Compiler | GNU Compiler | |
|---|---|---|---|
| Fortran 77 |
|
|
|
| Fortran 90 |
|
|
|
| Fortran 95 |
|
|
|
| C |
|
|
|
| C++ |
|
|
The Intel and GNU compilers will not output anything for a successful compilation. Also, the Intel compiler does not recognize the suffix ".f95".
Compiling MPI Programs
OpenMPI and Intel MPI (IMPI) are implementations of the Message-Passing Interface (MPI) standard. Libraries for these MPI implementations and compilers for C, C++, and Fortran are available on all clusters.
| Language | Header Files |
|---|---|
| Fortran 77 |
|
| Fortran 90 |
|
| Fortran 95 |
|
| C |
|
| C++ |
|
Here are a few sample programs using MPI:
To see the available MPI libraries:
$ module avail openmpi
$ module avail impi
| Language | Intel MPI | OpenMPI |
|---|---|---|
| Fortran 77 |
|
|
| Fortran 90 |
|
|
| Fortran 95 |
|
|
| C |
|
|
| C++ |
|
|
The Intel and GNU compilers will not output anything for a successful compilation. Also, the Intel compiler does not recognize the suffix ".f95".
Here is some more documentation from other sources on the MPI libraries:
Compiling OpenMP Programs
All compilers installed on Brown include OpenMP functionality for C, C++, and Fortran. An OpenMP program is a single process that takes advantage of a multi-core processor and its shared memory to achieve a form of parallel computing called multithreading. It distributes the work of a process over processor cores in a single compute node without the need for MPI communications.
| Language | Header Files |
|---|---|
| Fortran 77 |
|
| Fortran 90 |
|
| Fortran 95 |
|
| C |
|
| C++ |
|
Sample programs illustrate task parallelism of OpenMP:
A sample program illustrates loop-level (data) parallelism of OpenMP:
To load a compiler, enter one of the following:
$ module load intel
$ module load gcc
| Language | Intel Compiler | GNU Compiler |
|---|---|---|
| Fortran 77 |
|
|
| Fortran 90 |
|
|
| Fortran 95 |
|
|
| C |
|
|
| C++ |
|
|
The Intel and GNU compilers will not output anything for a successful compilation. Also, the Intel compiler does not recognize the suffix ".f95".
Here is some more documentation from other sources on OpenMP:
Frequently Asked Questions
Some common questions, errors, and problems are categorized below. Click the Expand Topics link in the upper right to see all entries at once. You can also use the search box above to search the user guide for any issues you are seeing.About Gautschi
Frequently asked questions about Gautschi.
Can you remove me from the Gautschi mailing list?
Your subscription in the Gautschi mailing list is tied to your account on Gautschi. If you are no longer using your account on Gautschi, your account can be deleted from the My Accounts page. Hover over the resource you wish to remove yourself from and click the red 'X' button. Your account and mailing list subscription will be removed overnight. Be sure to make a copy of any data you wish to keep first.
How is Gautschi different than other Community Clusters?
Gautschi differs from the previous Community Clusters in many significant aspects:
- Gautschi has a unique sales protocol. While previous clusters were available for sale based on compute resources, Gautschi is sold based on compute resource hours.
- Gautschi has eight NVIDIA H100 GPUs, each with 80 GB of GPU memory.
Do I need to do anything to my firewall to access Gautschi?
No firewall changes are needed to access Gautschi. However, to access data through Network Drives (i.e., CIFS, "Z: Drive"), you must be on a Purdue campus network or connected through VPN.
Does Gautschi have the same home directory as other clusters?
The Gautschi home directory and its contents are exclusive to Gautschi cluster front-end hosts and compute nodes. This home directory is not available on other RCAC machines but Gautschi. There is no automatic copying or synchronization between home directories.
At your discretion you can manually copy all or parts of your main research computing home to Gautschi using one of the suggested methods.
If you plan to use hsi or htar commands to access Fortress tape archive from Gautschi, please see also the keytab generation question for a temporary workaround to a potential caveat, while a permanent mitigation is being developed.
How should I launch common GUI applications on Gautschi?
When our users access Gautschi via ThinLinc, they can follow these steps to launch an interactive jobs.
- In the upper left corner, the user can click on Applications, then Cluster Software, where multiple software options are listed with interactive SLURM jobs.
- The GUI launcher starts with a window that prompts the user to select the desired version of the software to launch and guides them through the job submission process.
- The GUI launcher also makes it easy for users to view available accounts, the maximum wall times for each account, and the available computing resources with multiple help options at the bottom.
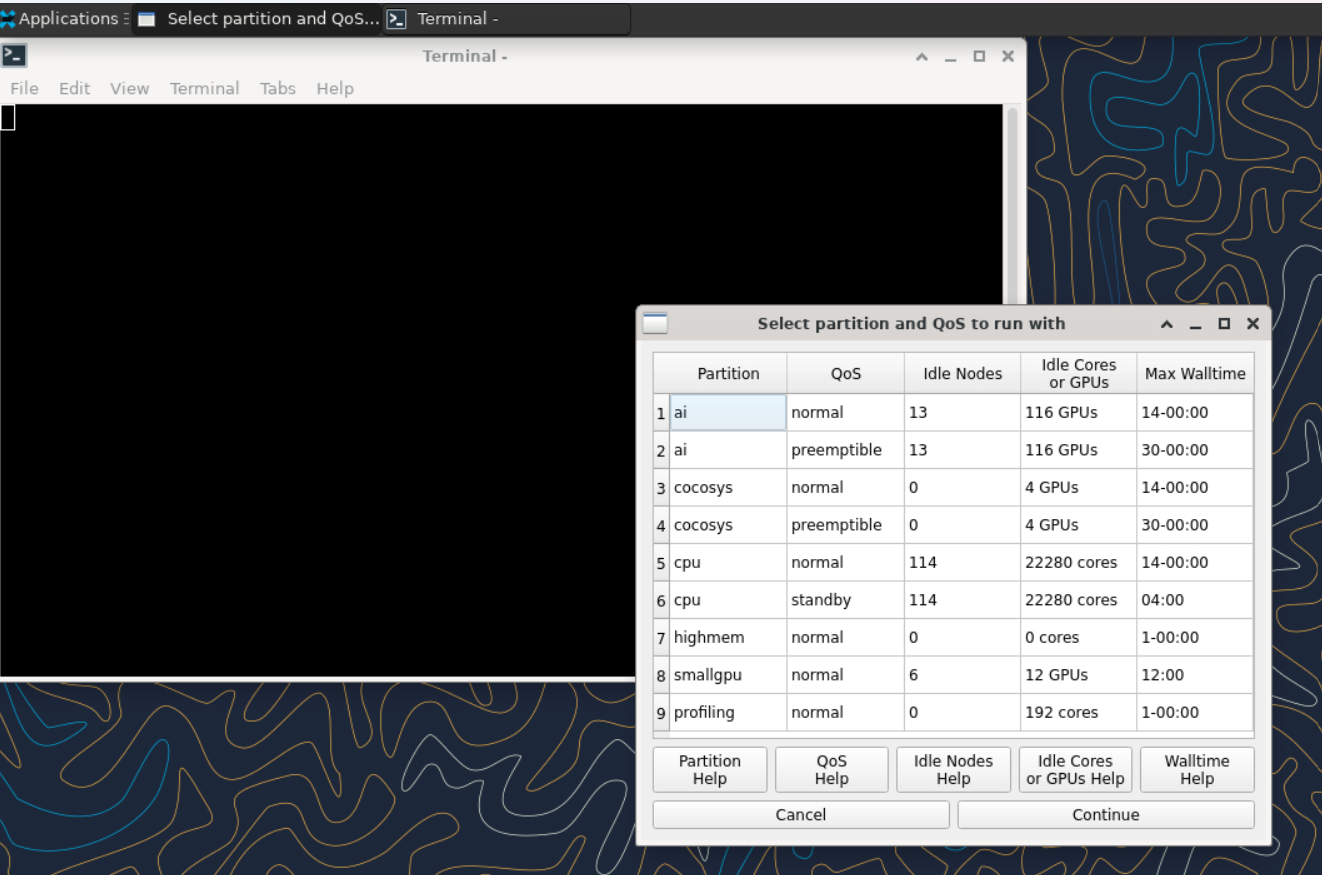
- After choosing the partition, the user is prompted to provide the computing resource they need for their job to run while adjusting the memory for them automatically.
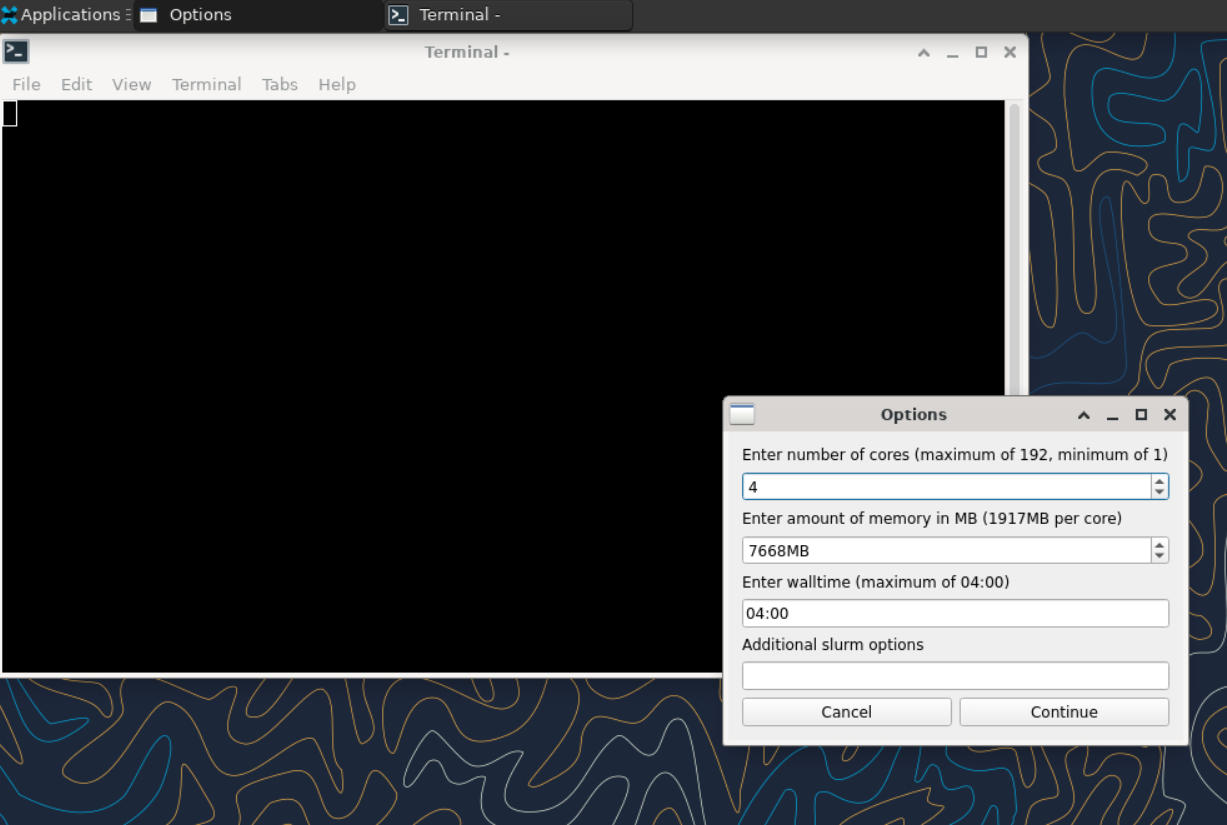
- After requesting the resources the job will be submitted and waiting for slurm to allocate the computing resources.
Software
Frequently asked questions about software.
Cannot use pip after loading ml-toolkit modules
Neither pip nor ml-toolkit are available on Gautschi, although they are available on the other community clusters.
Gateway (Open OnDemand)
Gautschi's Gateway is an open-source HPC portal developed by the Ohio Supercomputing Center. Open OnDemand allows one to interact with HPC resources through a web browser and easily manage files, submit jobs, and interact with graphical applications directly in a browser, all with no software to install. Gautschi has an instance of OnDemand available that can be accessed via gateway.gautschi.rcac.purdue.edu.
Link to section 'Logging In' of 'Gateway (Open OnDemand)' Logging In
To log into Gateway:
- Navigate to gateway.gautschi.rcac.purdue.edu
- Log in using your Career account username and Purdue Login Duo client.
On the splash page you will see a quota usage report. If you are over 90% on any of your quotas a warning will be displayed. This information will update every 10-15 minutes while you are active on Gateway.
Link to section 'Apps' of 'Gateway (Open OnDemand)' Apps
There are a number of built-in apps in Gateway that can be accessed from the top menu bar. Below are links to documentation on each app.
Files
The Files app will let you access your files in your Home Directory, Scratch, and Data Depot spaces. The app lets you manage create, manage, and delete files and directories from your web browser. Navigate by double clicking on folders in the file explorer or by using the file tree on the left.

On the top row, there are buttons to:
- Go To: directly input a directory to navigate to
- Open in Terminal: launches the Shell app and navigates you to the current directory in the terminal
- New File: creates a new, empty file
- New Dir: creates a new, empty directory
- Upload: upload a file from your computer
Note: File uploads from your browser are limited to 100 GB per file. Be mindful that uploads over a few gigabytes may be unreliable through your browser, especially from off-campus connections. For very large files or off-campus transfers alternative methods such as Globus are highly recommended.
The second row of buttons lets you perform typical file management operations. The Edit button will open files in a fully fledged browser based text editor - it features syntax highlighting and vim and Emacs key bindings.

Jobs
There are two apps under the Jobs apps: Active Jobs and Job Composer. These are detailed below.
Link to section 'Active Jobs' of 'Jobs' Active Jobs
This shows you active SLURM jobs currently on the cluster. The default view will show you your current jobs, similar to squeue -u rices. Using the button labeled "Your Jobs" in the upper right allows you to select different filters by queue (account). All accounts output by slist will appear for you here. Using the arrow on the left hand side will expand the full job details.

Link to section 'Job Composer' of 'Jobs' Job Composer
The Job Composer app allows you to create and submit jobs to the cluster. You can select from pre-defined templates (most of these are taken from the User Guide examples) or you can create your own templates for frequently used workflows.
Link to section 'Creating Job from Existing Template' of 'Jobs' Creating Job from Existing Template
Click "New Job" menu, then select "From Template":

Then select from one of the available templates.

Click 'Create New Job' in second pane.

Your new job should be selected in your list of jobs. In the 'Submit Script' pane you can see the job script that was generated with an 'Open Editor' link to open the script in the built-in editor. Open the file in the editor and edit the script as necessary. By default the job will specify standby queue - this should be changed as appropriate, along with the node and walltime requests.

When you are finished with editing the job and are ready to submit, click the green 'Submit' button at the top of the job list. You can monitor progress from here or from the Active Jobs app. Once completed, you should see the output files appear:

Clicking on one of the output files will open it in the file editor for your viewing.
Link to section 'Creating New Template' of 'Jobs' Creating New Template
First, prepare a template directory containing a template submission script along with any input files. Then, to import the job into the Job Composer app, click the 'Create New Template' button. Fill in the directory containing your template job script and files in the first box. Give it an appropriate name and notes.

This template will now appear in your list of templates to choose from when composing jobs. You can now go create and submit a job from this new template.
Cluster Tools
The Cluster Tools menu contains cluster utilities. At the moment, only a terminal app is provided. Additional apps may be developed and provided in the future.
Link to section 'Shell Access' of 'Cluster Tools' Shell Access
Launching the shell app will provide you with a web-based terminal session on the cluster front-end. This is equivalent to using a standalone SSH client to connect to gautschi.rcac.purdue.edu where you are connected to one several front-ends. The normal acceptable front-end use policy applies to access through the web-app. X11 Forwarding is not supported. Use of one of the interactive apps is recommended for graphical applications.
Interactive Apps
There are several interactive apps available through Gateway that can be accessed through the Interactive Apps dropdown menu. These apps are provided with a basic node and software configuration as a 'quick-launch' option to get your work up and running quickly. For simplicity, minimal options are provided - these apps are not intended for complex configuration/customization scenarios.
After you a submit an interactive app to the queue, Gateway will track and manage the session. Once it starts, you may connect and disconnect from the session in your browser, leaving the job running while you log out of your browser.
Each of the available apps are documented through the following links.
Compute Node Desktop
The Compute Node Desktop app will launch a graphical desktop session on a compute node. This is similar to using Thinlinc, however, this gives you a desktop directly on a compute node instead on a front-end. This app is useful if you have a custom application or application not directly available as an interactive app you would like to run inside Gateway.
To launch a desktop session on a compute node, select the Gautschi Compute Desktop app. From the submit form, select from the available options - the queue to which you wish to submit and the number of wallclock hours you wish to have job running. There is also a checkbox that enable a notification to your email when the job starts.
After the interactive job is submitted you will be taken to your list of active interactive app sessions. You can monitor the status of the job from here until it starts, or if you enabled the email notification, watch your Purdue email for the notification the job has started.
Once it is indicated the job has started you can connect to the desktop with the "Launch noVNC in New Tab" button. The session will be terminated after the wallclock hours you specified have elapsed or you terminate the session early with the "Delete" button from the list of sessions. Deleting the session when you are finished will free up queue resources for your lab mates and other users on the system.
MATLAB
The MATLAB app will launch a MATLAB session on a compute node and allow you to connect directly to it in a web browser.
To launch a MATLAB session on a compute node, select the MATLAB app. From the submit form, select from the available options - the version of MATLAB you are interested in running, the queue to which you wish to submit, and the number of wallclock hours you wish to have job running. There is also a checkbox that enable a notification to your email when the job starts.
After the interactive job is submitted you will be taken to your list of active interactive app sessions. You can monitor the status of the job from here until it starts, or if you enabled the email notification, watch your Purdue email for the notification the job has started.
Once it is indicated the job has started you can connect to the desktop with the "Launch noVNC in New Tab" button. The session will be terminated after the wallclock hours you specified have elapsed or you terminate the session early with the "Delete" button from the list of sessions. Deleting the session when you are finished will free up queue resources for your lab mates and other users on the system.
Jupyter Notebook - Deep Neural Networks Demo (GPU)
The Notebook app will launch a Notebook session on a compute node and allow you to connect directly to it in a web browser. It can be used to run GPU applications such as Tensorflow and Keras. Below is a demo of this to get you started.
- Download the demo notebook to your computer.
- Launch a Notebook session from the Gateway Interactive Apps menu:
- Select the queue to which you wish to submit and enter the number of wallclock hours you require. Your notebook will be terminated after this number of hours elapses.
- Click Launch.
- Wait for your interactive session to change to Running state. This may take some time depending on how busy the queue and system is.
- Click on 'Connect to Jupyter' once the button appears.
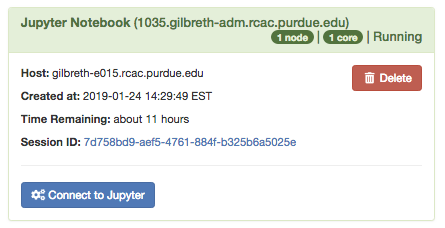
- Once in Jupyter, select 'Upload' in the upper right corner. You may wish to create a folder or change into a different directory to put the demo notebook first.
- Select the demo notebook file you downloaded earlier. Click the blue Upload button to complete the upload. Then click the
dnn.ipynbitem from the file list to launch the notebook. - You should now have the notebook loaded and you should be able to re-execute the code cells, or modify them to your needs.
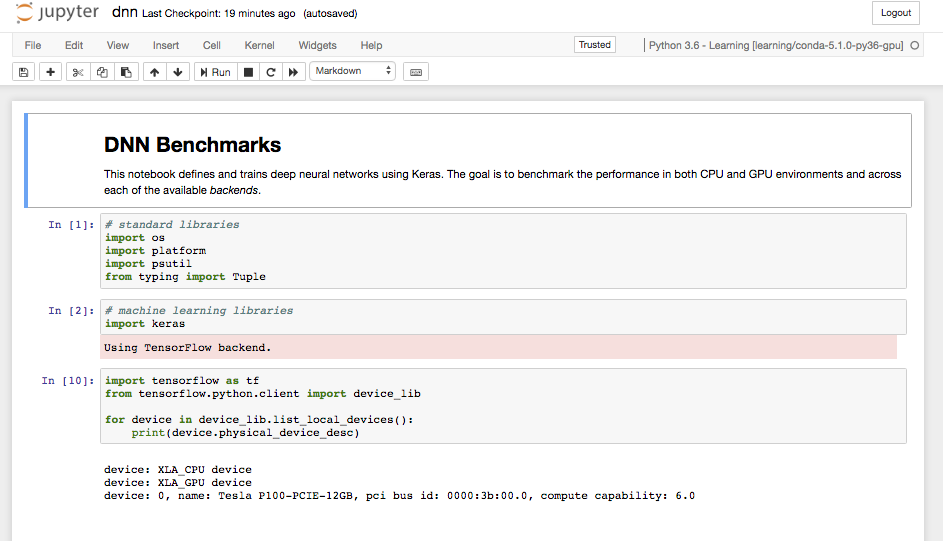
Jupyter Notebook
The Notebook app will launch a Notebook session on a compute node and allow you to connect directly to it in a web browser.
To launch a Notebook session on a compute node, select the Notebook app. From the submit form, select from the available options:
- Queue: This is a dropdown menu from which you can select a queue from all of the queues to which you have permission to submit.
- Walltime: This is a field which expects a number and represents how many hours you want to keep the session running. Note that this value should not exceed the maximum value given next to the selected queue name from the queue dropdown menu.
- Number of Cores/GPUs: This is a field which expects a number and represents the number of your resources your session is requesting. Note that the amount of memory allocated for your session is proportional to the number of cores or GPUs that you request for your job, so if your session is running out of memory, consider increasing this value.
- Use Jupyter Lab: This is a checkbox which, when checked, will run Jupyter Lab instead of Jupyter Notebook. Both of these applications are interfaces to Jupyter, and you can launch Jupyter notebooks from within Jupyter Lab. Jupyter Notebook is more "barebones" while Jupyter Lab has additional features such as the ability to interact with additional file types.
- E-mail Notice: This is a checkbox which, when checked, will send you an e-mail notification to your Purdue e-mail that your session is ready when the scheduler has found resources to dedicate to your session.
After the interactive job is submitted you will be taken to your list of active interactive app sessions. You can monitor the status of the job from here until it starts, or if you enabled the email notification, watch your Purdue email for the notification the job has started.
Once it is indicated the job has started you can connect to the desktop with the "Connect to Jupyter" button. Once connected, you can create new notebooks, selecting the currently available Anaconda versions available as modules, and any personally created Notebook kernels.
Often times you may want to use one of your existing Anaconda environments within your Jupyter session to use libraries specific to your workflow. In order to do so, you must ensure that the Anaconda environment you want to use contains the Python packages "IPyKernel" and "IPython" which are packages that are required by Jupyter. When you create a Jupyter session, Open OnDemand will check through your existing Anaconda environments and create a Jupyter kernel for any Anaconda environment that contains these two packages, and you will be able to select to use that kernel from within the application.
The session will be terminated after the number of hours you specified have elapsed or you terminate the session early with the "Delete" button from the list of sessions. Deleting the session when you are finished will free up queue resources for your lab mates and other users on the system.
RStudio Server
The RStudio app will launch a RStudio session on a compute node and allow you to connect directly to it in a web browser.
To launch a RStudio session on a compute node, select the RStudio app. From the submit form, select from the available options - the queue to which you wish to submit, and the number of wallclock hours you wish to have job running. There is also a checkbox that enable a notification to your email when the job starts.
After the interactive job is submitted you will be taken to your list of active interactive app sessions. You can monitor the status of the job from here until it starts, or if you enabled the email notification, watch your Purdue email for the notification the job has started.
Once it is indicated the job has started you can connect to the desktop with the "Connect to RStudio Server" button. The session will be terminated after the wallclock hours you specified have elapsed or you terminate the session early with the "Delete" button from the list of sessions. Deleting the session when you are finished will free up queue resources for your lab mates and other users on the system.
Windows Desktop
The Windows Desktop app will launch a Windows desktop session on a compute node. This is similar to using the Windows menu launcher through Thinlinc, however, this gives you a Windows desktop directly on a compute node instead on a front-end.
To launch a Windows session on a compute node, select the Windows Desktop app. From the submit form, select from the available options - choose from the basic Windows configuration or the GIS configured image, the queue to which you wish to submit, and the number of wallclock hours you wish to have job running. There is also a checkbox that enable a notification to your email when the job starts.
This will create a file in your scratch space called windows-base.qcow2 or windows-gis.qcow2. If the file already exists, the existing image will be restarted. You can delete or rename the image at any time through the Files App to generate a fresh image. You can only have one instance of the image running at a time or corruption will occur. There are lock files to prevent this, but be mindful of this restriction. It is also recommended you make periodic backups of the image if you are making any modifications to it.
After the interactive job is submitted you will be taken to your list of active interactive app sessions. You can monitor the status of the job from here until it starts, or if you enabled the email notification, watch your Purdue email for the notification the job has started.
Once it is indicated the job has started you can connect to the desktop with the "Launch noVNC in New Tab" button. The session will be terminated after the wallclock hours you specified have elapsed or you terminate the session early with the "Delete" button from the list of sessions. Deleting the session when you are finished will free up queue resources for your lab mates and other users on the system.
Datasets
Please refer to our Federated Datasets Documentation website for up-to-date datasets on Anvil and instructions on how to use them.
