Applications
Link to section 'AMD' of 'Applications' AMD
Link to section 'Audio/Visualization' of 'Applications' Audio/Visualization
Link to section 'Bioinformatics' of 'Applications' Bioinformatics
Link to section 'Climate' of 'Applications' Climate
Link to section 'Computational chemistry' of 'Applications' Computational chemistry
Link to section 'Container' of 'Applications' Container
Link to section 'Electrical engineering' of 'Applications' Electrical engineering
Link to section 'Fluid dynamics' of 'Applications' Fluid dynamics
Link to section 'Geospatial tools' of 'Applications' Geospatial tools
Link to section 'Libraries' of 'Applications' Libraries
- arpack-ng
- blis
- boost
- eigen
- fftw
- gmp
- ghostscript
- gsl
- hdf5
- hdf
- intel-mkl
- intel-oneapi-mkl
- intel-oneapi-tbb
- intel-rt
- libfabric
- libflame
- libiconv
- libmesh
- libpng
- libszip
- libtiff
- libv8
- libx11
- libxml2
- libxp
- libxscrnsaver
- libxslt
- mesa-glu
- mpfr
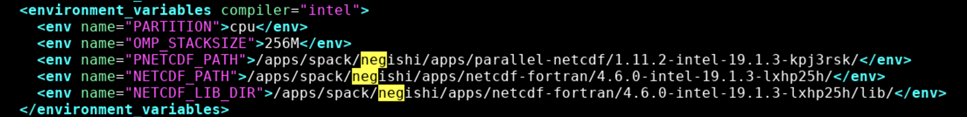
- netcdf-c
- netcdf-cxx4
- netcdf-fortran
- netcdf
- netlib-lapack
- oclfpga
- openblas
- parallel-netcdf
- petsc
- rocm
- sqlite
- swig
- ucx
- udunits2
- udunits
- util-linux-uuid
- xcb-util-image
- xcb-util-keysyms
- xcb-util-renderutil
- xcb-util-wm
- zlib
- zstd
Link to section 'Material science' of 'Applications' Material science
Link to section 'Mathematical/Statistics' of 'Applications' Mathematical/Statistics
Link to section 'ML toolkit' of 'Applications' ML toolkit
Link to section 'NVIDIA' of 'Applications' NVIDIA
Link to section 'Physics' of 'Applications' Physics
Link to section 'Programming languages' of 'Applications' Programming languages
Link to section 'System' of 'Applications' System
Link to section 'Text Editors' of 'Applications' Text Editors
Link to section 'Tools/Utilities' of 'Applications' Tools/Utilities
Link to section 'Workflow automation' of 'Applications' Workflow automation
abaqus
Link to section 'Description' of 'abaqus' Description
Abaqus is a software suite for Finite Element Analysis (FEA) developed by Dassault Systèmes.
Link to section 'Versions' of 'abaqus' Versions
- Bell: 2019, 2020, 2021, 2022, 2023
- Scholar: 2024
- Gilbreth: 2017, 2018, 2019, 2020, 2021, 2022
- Negishi: 2022, 2023, 2024
Link to section 'Module' of 'abaqus' Module
You can load the modules by:
module load abaqus
amber
Link to section 'Description' of 'amber' Description
AMBER (Assisted Model Building with Energy Refinement) is a package of molecular simulation programs.
Link to section 'Versions' of 'amber' Versions
- Bell: 16
- Gilbreth: 16
- Negishi: 20
- Anvil: 20
Link to section 'Module' of 'amber' Module
You can load the modules by:
module load amber
amdblis
Link to section 'Description' of 'amdblis' Description
AMD Optimized BLIS. BLIS is a portable software framework for instantiating high-performance BLAS-like dense linear algebra libraries.
Link to section 'Versions' of 'amdblis' Versions
- Anvil: 3.0
Link to section 'Module' of 'amdblis' Module
You can load the modules by:
module load amdblis
amdfftw
Link to section 'Description' of 'amdfftw' Description
FFTW AMD Optimized version is a comprehensive collection of fast C routines for computing the Discrete Fourier Transform DFT and various special cases thereof.
Link to section 'Versions' of 'amdfftw' Versions
- Anvil: 3.0
Link to section 'Module' of 'amdfftw' Module
You can load the modules by:
module load amdfftw
amdlibflame
Link to section 'Description' of 'amdlibflame' Description
libFLAME AMD Optimized version is a portable library for dense matrix computations, providing much of the functionality present in Linear Algebra Package LAPACK. It includes a compatibility layer, FLAPACK, which includes complete LAPACK implementation.
Link to section 'Versions' of 'amdlibflame' Versions
- Anvil: 3.0
Link to section 'Module' of 'amdlibflame' Module
You can load the modules by:
module load amdlibflame
amdlibm
Link to section 'Description' of 'amdlibm' Description
AMD LibM is a software library containing a collection of basic math functions optimized for x86-64 processor-based machines. It provides many routines from the list of standard C99 math functions. Applications can link into AMD LibM library and invoke math functions instead of compilers math functions for better accuracy and performance.
Link to section 'Versions' of 'amdlibm' Versions
- Anvil: 3.0
Link to section 'Module' of 'amdlibm' Module
You can load the modules by:
module load amdlibm
amdscalapack
Link to section 'Description' of 'amdscalapack' Description
ScaLAPACK is a library of high-performance linear algebra routines for parallel distributed memory machines. It depends on external libraries including BLAS and LAPACK for Linear Algebra computations.
Link to section 'Versions' of 'amdscalapack' Versions
- Anvil: 3.0
Link to section 'Module' of 'amdscalapack' Module
You can load the modules by:
module load amdscalapack
anaconda
Link to section 'Description' of 'anaconda' Description
Anaconda is a free and open-source distribution of the Python and R programming languages for scientific computing, that aims to simplify package management and deployment.
Link to section 'Versions' of 'anaconda' Versions
- Bell: 2019.10-py27, 2020.02-py37, 2020.11-py38, 2024.02-py311
- Gilbreth: 2019.10-py27, 2020.02-py37, 2020.11-py38, 2024.02-py311, 5.1.0-py27, 5.1.0-py36, 5.3.1-py27, 5.3.1-py37
- Negishi: 2021.05-py38, 2022.10-py39, 2024.02-py311
- Anvil: 2021.05-py38, 2024.02-py311
Link to section 'Module' of 'anaconda' Module
You can load the modules by:
module load anaconda
ansys
Link to section 'Description' of 'ansys' Description
Ansys is a CAE/multiphysics engineering simulation software that utilizes finite element analysis for numerically solving a wide variety of mechanical problems. The software contains a list of packages and can simulate many structural properties such as strength, toughness, elasticity, thermal expansion, fluid dynamics as well as acoustic and electromagnetic attributes.
Link to section 'Versions' of 'ansys' Versions
- Bell: 2019R3, 2020R1, 2021R2, 2022R1, 2022R2, 2023R1, 2023R2
- Scholar: 2022R1
- Gilbreth: 19.2, 2019R3, 2020R1, 2021R2, 2022R1, 2023R1, 2023R2
- Negishi: 2022R2, 2023R1, 2023R2, 2024R2
- Gautschi: 2024R2
Link to section 'Ansys Licensing' of 'ansys' Ansys Licensing
The Ansys licensing on our community clusters is maintained by Purdue ECN group. There are two types of licenses: teaching and research. For more information, please refer to ECN Ansys licensing page. If you are interested in purchasing your own research license, please send email to software@ecn.purdue.edu.
Link to section 'Ansys Workflow' of 'ansys' Ansys Workflow
Ansys software consists of several sub-packages such as Workbench and Fluent. Most simulations are performed using the Ansys Workbench console, a GUI interface to manage and edit the simulation workflow. It requires X11 forwarding for remote display so a SSH client software with X11 support or a remote desktop portal is required. Please see Logging In section for more details. To ensure preferred performance, ThinLinc remote desktop connection is highly recommended.
Typically users break down larger structures into small components in geometry with each of them modeled and tested individually. A user may start by defining the dimensions of an object, adding weight, pressure, temperature, and other physical properties.
Ansys Fluent is a computational fluid dynamics (CFD) simulation software known for its advanced physics modeling capabilities and accuracy. Fluent offers unparalleled analysis capabilities and provides all the tools needed to design and optimize new equipment and to troubleshoot existing installations.
In the following sections, we provide step-by-step instructions to lead you through the process of using Fluent. We will create a classical elbow pipe model and simulate the fluid dynamics when water flows through the pipe. The project files have been generated and can be downloaded via fluent_tutorial.zip.
Link to section 'Loading Ansys Module' of 'ansys' Loading Ansys Module
Different versions of Ansys are installed on the clusters and can be listed with module spider or module avail command in the terminal.
$ module avail ansys/
---------------------- Core Applications -----------------------------
ansys/2019R3 ansys/2020R1 ansys/2021R2 ansys/2022R1 (D)
Before launching Ansys Workbench, a specific version of Ansys module needs to be loaded. For example, you can module load ansys/2021R2 to use the latest Ansys 2021R2. If no version is specified, the default module -> (D) (ansys/2022R1 in this case) will be loaded. You can also check the loaded modules with module list command.
Link to section 'Launching Ansys Workbench' of 'ansys' Launching Ansys Workbench
Open a terminal, enter rcac-runwb2 to launch Ansys Workbench.
You can also use runwb2 to launch Ansys Workbench. The main difference between runwb2and rcac-runwb2 is that the latter sets the project folder to be in your scratch space. Ansys has an known bug that it might crash when the project folder is set to $HOME on our systems.
ansysem
Link to section 'Description' of 'ansysem' Description
This module enables the use of ANSYSEM, a popular Electromechanical application.
Link to section 'Versions' of 'ansysem' Versions
- Bell: 2020r1, 2021r2
- Scholar: 2021r2
Link to section 'Module' of 'ansysem' Module
You can load the modules by:
module load ansysem
aocl
Link to section 'Description' of 'aocl' Description
AOCL are a set of numerical libraries tuned specifically for AMD EPYCTM processor family. They have a simple interface to take advantage of the latest hardware innovations.
Link to section 'Versions' of 'aocl' Versions
- Bell: 2.1
Link to section 'Module' of 'aocl' Module
You can load the modules by:
module load aocl
arpack-ng
Link to section 'Description' of 'arpack-ng' Description
ARPACK-NG is a collection of Fortran77 subroutines designed to solve large scale eigenvalue problems.
Link to section 'Versions' of 'arpack-ng' Versions
- Negishi: 3.8.0
- Anvil: 3.8.0
Link to section 'Module' of 'arpack-ng' Module
You can load the modules by:
module load arpack-ng
aws-cli
Link to section 'Description' of 'aws-cli' Description
The AWS Command Line Interface CLI is a unified tool to manage your AWS services from command line.
Link to section 'Versions' of 'aws-cli' Versions
- Bell: 2.4.15
- Scholar: 2.4.15
- Gilbreth: 2.4.15
- Negishi: 2.9.7
- Anvil: 2.4.15
Link to section 'Module' of 'aws-cli' Module
You can load the modules by:
module load aws-cli
bamtools
Link to section 'Description' of 'bamtools' Description
C++ API & command-line toolkit for working with BAM data.
Link to section 'Versions' of 'bamtools' Versions
- Anvil: 2.5.1, 2.5.2
- Bell: 2.5.1
- Negishi: 2.5.1
- Scholar: 2.5.1
- Gautschi: 2.5.1
Link to section 'Commands' of 'bamtools' Commands
- bamtools
Link to section 'Module' of 'bamtools' Module
You can load the modules by:
module load bamtools
Link to section 'Example job' of 'bamtools' Example job
To run bamtools our our clusters:
#!/bin/bash
#SBATCH -A Allocation # Allocation name
#SBATCH -t 20:00:00
#SBATCH -N 1
#SBATCH -n 24
#SBATCH -p PartitionName
#SBATCH --job-name=bamtools
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load bamtools
bamtools convert -format fastq -in in.bam -out out.fastq
bbftp
Link to section 'Description' of 'bbftp' Description
bbFTP is a file transfer software. It implements its own transfer protocol, which is optimized for large files (larger than 2GB) and secure as it does not read the password in a file and encrypts the connection information.
Link to section 'Versions' of 'bbftp' Versions
- Bell: 3.2.1
Link to section 'Module' of 'bbftp' Module
You can load the modules by:
module load bbftp
beagle
Link to section 'Description' of 'beagle' Description
Beagle is a software package for phasing genotypes and for imputing ungenotyped markers.
Link to section 'Versions' of 'beagle' Versions
- Anvil: 5.1
- Bell: 5.1
- Negishi: 5.1
- Scholar: 5.1_24Aug19.3e8
- Gautschi: 5.1_24Aug19.3e8
Link to section 'Commands' of 'beagle' Commands
- beagle
Link to section 'Module' of 'beagle' Module
You can load the modules by:
module load beagle
Link to section 'Example job' of 'beagle' Example job
To run Beagle on our clusters:
#!/bin/bash
#SBATCH -A myAllocation
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 1
#SBATCH --job-name=beagle
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load beagle
beagle gt=test.vcf.gz out=test.out
beast2
Link to section 'Description' of 'beast2' Description
BEAST is a cross-platform program for Bayesian inference using MCMC of molecular sequences. It is entirely orientated towards rooted, time-measured phylogenies inferred using strict or relaxed molecular clock models. It can be used as a method of reconstructing phylogenies but is also a framework for testing evolutionary hypotheses without conditioning on a single tree topology.
Link to section 'Versions' of 'beast2' Versions
- Anvil: 2.6.3, 2.6.4, 2.6.6
- Bell: 2.6.3, 2.6.4, 2.6.6
- Negishi: 2.6.3, 2.6.4, 2.6.6
- Scholar: 2.6.3, 2.6.4, 2.6.6
- Gautschi: 2.6.3, 2.6.4, 2.6.6
Link to section 'Commands' of 'beast2' Commands
- applauncher
- beast
- beauti
- densitree
- loganalyser
- logcombiner
- packagemanager
- treeannotator
Link to section 'Module' of 'beast2' Module
You can load the modules by:
module load beast2
Link to section 'Example job' of 'beast2' Example job
To run BEAST 2 on our clusters:
#!/bin/bash
#SBATCH -A myQueue
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 4
#SBATCH --job-name=beast2
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load beast2
beast -threads 4 -prefix input input.xml
bismark
Link to section 'Description' of 'bismark' Description
A tool to map bisulfite converted sequence reads and determine cytosine methylation states
Link to section 'Versions' of 'bismark' Versions
- Anvil: 0.23.0, 0.24.0
- Bell: 0.23.0, 0.24.0
- Negishi: 0.23.0, 0.24.0
- Scholar: 0.23.0, 0.24.0
- Gautschi: 0.23.0, 0.24.0
Link to section 'Commands' of 'bismark' Commands
- bam2nuc
- bismark
- bismark2bedGraph
- bismark2report
- bismark2summary
- bismark_genome_preparation
- bismark_methylation_extractor
- coverage2cytosine
- deduplicate_bismark
- filter_non_conversion
- NOMe_filtering
Link to section 'Module' of 'bismark' Module
You can load the modules by:
module load bismark
Link to section 'Example job' of 'bismark' Example job
To run Bismark on our clusters:
#!/bin/bash
#SBATCH -A myAllocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 12
#SBATCH --job-name=bismark
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load bismark
bismark_genome_preparation --bowtie2 data/ref_genome
bismark --multicore 12 --genome data/ref_genome seq.fastq
blast-plus
Link to section 'Description' of 'blast-plus' Description
Basic Local Alignment Search Tool. BLAST finds regions of similarity between biological sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance.
Link to section 'Versions' of 'blast-plus' Versions
- Anvil: 2.12.0
Link to section 'Module' of 'blast-plus' Module
You can load the modules by:
module load blast-plus
Link to section 'BLAST Databases' of 'blast-plus' BLAST Databases
Local copies of the blast dabase can be found in the directory /anvil/datasets/ncbi/blast/latest. The environment varialbe BLASTDB was also set as /anvil/datasets/ncbi/blast/latest. If users want to use cdd_delta, env_nr, env_nt, nr, nt, pataa, patnt, pdbnt, refseq_protein, refseq_rna, swissprot, or tsa_nt databases, do not need to provide the database path. Instead, just use the format like this -db nr.
Link to section 'Example job' of 'blast-plus' Example job
To run bamtools our our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 20:00:00
#SBATCH -N 1
#SBATCH -n 24
#SBATCH -p PartitionName
#SBATCH --job-name=blast
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load blast-plus
blastp -query protein.fasta -db nr -out test_out -num_threads 4
blis
Link to section 'Description' of 'blis' Description
BLIS is a portable software framework for instantiating high-performance BLAS-like dense linear algebra libraries.
Link to section 'Versions' of 'blis' Versions
- Negishi: 0.8.1, 0.9.0
- Anvil: 0.8.1
Link to section 'Module' of 'blis' Module
You can load the modules by:
module load blis
boost
Link to section 'Description' of 'boost' Description
Boost provides free peer-reviewed portable C++ source libraries, emphasizing libraries that work well with the C++ Standard Library.
Link to section 'Versions' of 'boost' Versions
- Bell: 1.68.0, 1.70.0, 1.80.0
- Scholar: 1.85.0
- Gilbreth: 1.66.0, 1.70.0, 1.79.0
- Negishi: 1.79.0, 1.80.0
- Anvil: 1.74.0
- Gautschi: 1.85.0
Link to section 'Module' of 'boost' Module
You can load the modules by:
module load boost
bowtie2
Link to section 'Description' of 'bowtie2' Description
Bowtie 2 is an ultrafast and memory-efficient tool for aligning sequencing reads to long reference sequences
Link to section 'Versions' of 'bowtie2' Versions
- Anvil: 2.4.2, 2.5.1
- Bell: 2.4.2, 2.5.1
- Negishi: 2.4.2, 2.5.1
- Scholar: 2.4.2, 2.5.1
- Gautschi: 2.4.2, 2.5.1
Link to section 'Module' of 'bowtie2' Module
You can load the modules by:
module load bowtie2
Link to section 'Example job' of 'bowtie2' Example job
To run Bowtie 2 on our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 1
#SBATCH --job-name=bowtie2
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load bowtie2
bowtie2-build ref.fasta ref
bowtie2 -p 4 -x ref -1 input_1.fq -2 input_2.fq -S test.sam
bwa
Link to section 'Description' of 'bwa' Description
Burrow-Wheeler Aligner for pairwise alignment between DNA sequences.
Link to section 'Versions' of 'bwa' Versions
- Anvil: 0.7.17
- Bell: 0.7.17
- Negishi: 0.7.17
- Scholar: 0.7.17
- Gautschi: 0.7.17
Link to section 'Module' of 'bwa' Module
You can load the modules by:
module load bwa
Link to section 'Example job' of 'bwa' Example job
To run BWA on our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 1
#SBATCH --job-name=bwa
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load bwa
bwa index ref.fasta
bwa mem ref.fasta input.fq > test.sam
bzip2
Link to section 'Description' of 'bzip2' Description
bzip2 is a freely available, patent free high-quality data compressor. It typically compresses files to within 10% to 15% of the best available techniques the PPM family of statistical compressors, whilst being around twice as fast at compression and six times faster at decompression.
Link to section 'Versions' of 'bzip2' Versions
- Bell: 1.0.8
- Gilbreth: 1.0.8
- Negishi: 1.0.8
- Anvil: 1.0.8
Link to section 'Module' of 'bzip2' Module
You can load the modules by:
module load bzip2
caffe
Link to section 'Description' of 'caffe' Description
Caffe is a deep learning framework made with expression, speed, and modularity in mind.
Link to section 'Versions' of 'caffe' Versions
- Bell: 1.0
- Gilbreth: 1.0.0
Link to section 'Module' of 'caffe' Module
You can load the modules by:
module load learning
module load caffe
cdo
Link to section 'Description' of 'cdo' Description
CDO is a collection of command line Operators to manipulate and analyse Climate and NWP model Data.
Link to section 'Versions' of 'cdo' Versions
- Bell: 1.9.5
- Scholar: 2.3.0
- Gilbreth: 1.9.5
- Negishi: 1.9.9
- Anvil: 1.9.9
- Gautschi: 2.3.0
Link to section 'Module' of 'cdo' Module
You can load the modules by:
module load cdo
cmake
Link to section 'Description' of 'cmake' Description
A cross-platform, open-source build system. CMake is a family of tools designed to build, test and package software.
Link to section 'Versions' of 'cmake' Versions
- Bell: 3.18.2, 3.20.6, 3.23.1, 3.30.1
- Scholar: 3.26.5
- Gilbreth: 3.15.4, 3.20.6
- Negishi: 3.23.1, 3.24.3
- Anvil: 3.20.0
- Gautschi: 3.30.2, 3.30.2
Link to section 'Module' of 'cmake' Module
You can load the modules by:
module load cmake
cntk
Link to section 'Description' of 'cntk' Description
The Microsoft Cognitive Toolkit is a unified deep-learning toolkit that describes neural networks as a series of computational steps via a directed graph.
Link to section 'Versions' of 'cntk' Versions
- Gilbreth: 2.6
Link to section 'Module' of 'cntk' Module
You can load the modules by:
module load learning
module load cntk
comsol
Link to section 'Description' of 'comsol' Description
Comsol Multiphysics (previously named Femlab) is a modeling package for the simulation of any physical process you can describe with partial differential equations (PDEs).
Link to section 'Versions' of 'comsol' Versions
- Bell: 5.3a, 5.4, 5.5_b359, 5.6, 6.0, 6.1, 6.2
- Scholar: 6.2
- Negishi: 6.1, 6.2
- Gautschi: 6.2
Link to section 'Module' of 'comsol' Module
You can load the modules by:
module load comsol
cp2k
Link to section 'Description' of 'cp2k' Description
CP2K is a quantum chemistry and solid state physics software package that can perform atomistic simulations of solid state, liquid, molecular, periodic, material, crystal, and biological systems
Link to section 'Versions' of 'cp2k' Versions
- Negishi: 20210311, 2022.1, 8.2
- Anvil: 8.2
- Bell: 20210311, 8.2
Link to section 'Module' of 'cp2k' Module
You can load the modules by:
module load cp2k
cplex
Link to section 'Description' of 'cplex' Description
IBM ILOG CPLEX Optimizer's mathematical programming technology enables decision optimization for improving efficiency, reducing costs, and increasing profitability.
Link to section 'Versions' of 'cplex' Versions
- Bell: 12.8.0
Link to section 'Module' of 'cplex' Module
You can load the modules by:
module load cplex
cuda
Link to section 'Description' of 'cuda' Description
CUDA is a parallel computing platform and programming model invented by NVIDIA. It enables dramatic increases in computing performance by harnessing the power of the graphics processing unit (GPU).
Link to section 'Versions' of 'cuda' Versions
- Scholar: 12.1.0
- Gilbreth: 8.0.61, 9.0.176, 10.0.130, 10.2.89, 11.0.3, 11.2.0, 11.7.0, 12.1.1
- Anvil: 11.0.3, 11.2.2, 11.4.2, 12.0.1
- Gautschi: 12.6.0
Link to section 'Module' of 'cuda' Module
You can load the modules by:
module load cuda
Link to section 'Monitor Activity and Drivers' of 'cuda' Monitor Activity and Drivers
Users can check the available GPUs, their current usage, installed version of the nvidia drivers, and running processes with the command nvidia-smi. The output should look something like this:
User@gilbreth-fe00:~/cuda $ nvidia-smi
Sat May 27 23:26:14 2023
+-----------------------------------------------------------------------------+
| NVIDIA-SMI 515.48.07 Driver Version: 515.48.07 CUDA Version: 11.7 |
|-------------------------------+----------------------+----------------------+
| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|===============================+======================+======================|
| 0 NVIDIA A30 Off | 00000000:21:00.0 Off | 0 |
| N/A 29C P0 29W / 165W | 19802MiB / 24576MiB | 0% Default |
| | | Disabled |
+-------------------------------+----------------------+----------------------+
+-----------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=============================================================================|
| 0 N/A N/A 29152 C python 9107MiB |
| 0 N/A N/A 53947 C ...020.11-py38/GP/bin/python 2611MiB |
| 0 N/A N/A 71769 C ...020.11-py38/GP/bin/python 1241MiB |
| 0 N/A N/A 72821 C ...8/TorchGPU_env/bin/python 2657MiB |
| 0 N/A N/A 91986 C ...2-4/internal/bin/gdesmond 931MiB |
+-----------------------------------------------------------------------------+
We can see that the node gilbreth-fe00 is running driver version 515.48.07 and is compatible with CUDA version 11.7. We do not recommend users to run jobs on front end nodes, but here we can see there are three python processes and one gdesmond process.
Link to section 'Compile a CUDA code' of 'cuda' Compile a CUDA code
The below vectorAdd.cu is modified from the textbook Learn CUDA Programming.
#include<stdio.h>
#include<stdlib.h>
#define N 512
void host_add(int *a, int *b, int *c) {
for(int idx=0;idx<N;idx++)
c[idx] = a[idx] + b[idx];
}
//basically just fills the array with index.
void fill_array(int *data) {
for(int idx=0;idx<N;idx++)
data[idx] = idx;
}
void print_output(int *a, int *b, int*c) {
for(int idx=0;idx<N;idx++)
printf("\n %d + %d = %d", a[idx] , b[idx], c[idx]);
}
int main(void) {
int *a, *b, *c;
int size = N * sizeof(int);
// Alloc space for host copies of a, b, c and setup input values
a = (int *)malloc(size); fill_array(a);
b = (int *)malloc(size); fill_array(b);
c = (int *)malloc(size);
host_add(a,b,c);
print_output(a,b,c);
free(a); free(b); free(c);
return 0;
}
We can compile the CUDA code by the CUDA nvcc compiler:
nvcc -o vector_addition vector_addition.cu
Link to section 'Example job script' of 'cuda' Example job script
#!/bin/bash
#SBATCH -A XXX
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=20
#SBATCH --cpus-per-task=1
#SBATCH --gpus-per-node=1
#SBATCH --time 1:00:00
module purge
module load gcc/XXX
module load cuda/XXX
#compile the vector_addition.cu file
nvcc -o vector_addition vector_addition.cu
#runs the vector_addition program
./vector_addition
cudnn
Link to section 'Description' of 'cudnn' Description
cuDNN is a deep neural network library from Nvidia that provides a highly tuned implementation of many functions commonly used in deep machine learning applications.
Link to section 'Versions' of 'cudnn' Versions
- Gilbreth: cuda-8.0_6.0, cuda-8.0_7.1, cuda-9.0_7.3, cuda-9.0_7.4, cuda-10.0_7.5, cuda-10.2_8.0, cuda-11.0_8.0, cuda-11.2_8.1, cuda-11.7_8.6, cuda-12.1_8.9
- Anvil: cuda-11.0_8.0, cuda-11.2_8.1, cuda-11.4_8.2, cuda-12.0_8.8
Link to section 'Module' of 'cudnn' Module
You can load the modules by:
module load cudnn
cue-login-env
Link to section 'Description' of 'cue-login-env' Description
XSEDE Common User Environment Variables for Anvil. Load this module to have XSEDE Common User Environment variables defined for your shell session or job on Anvil. See detailed description at https://www.ideals.illinois.edu/bitstream/handle/2142/75910/XSEDE-CUE-Variable-Definitions-v1.1.pdf
Link to section 'Versions' of 'cue-login-env' Versions
- Anvil: 1.1
Link to section 'Module' of 'cue-login-env' Module
You can load the modules by:
module load cue-login-env
cufflinks
Link to section 'Description' of 'cufflinks' Description
Cufflinks assembles transcripts, estimates their abundances, and tests for differential expression and regulation in RNA-Seq samples.
Link to section 'Versions' of 'cufflinks' Versions
- Anvil: 2.2.1
- Bell: 2.2.1
- Negishi: 2.2.1
- Scholar: 2.2.1
- Gautschi: 2.2.1
Link to section 'Module' of 'cufflinks' Module
You can load the modules by:
module load cufflinks
Link to section 'Example job' of 'cufflinks' Example job
To run Cufflinks on our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 8
#SBATCH --job-name=cufflinks
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load cufflinks
cufflinks -p 8 -G transcript.gtf --library-type fr-unstranded -o cufflinks_output tophat_out/accepted_hits.bam
curl
Link to section 'Description' of 'curl' Description
cURL is an open source command line tool and library for transferring data with URL syntax
Link to section 'Versions' of 'curl' Versions
- Bell: 7.63.0, 7.79.0, 8.7.1
- Scholar: 7.76.1
- Gilbreth: 7.79.0
- Negishi: 7.78.0, 7.85.0
- Anvil: 7.76.1
- Gautschi: 8.8.0
Link to section 'Module' of 'curl' Module
You can load the modules by:
module load curl
cutadapt
Link to section 'Description' of 'cutadapt' Description
Cutadapt finds and removes adapter sequences, primers, poly-A tails and other types of unwanted sequence from your high-throughput sequencing reads.
Link to section 'Versions' of 'cutadapt' Versions
- Anvil: 2.10, 3.4, 3.7
- Bell: 3.4, 3.7
- Negishi: 3.4, 3.7
- Scholar: 3.4, 3.7
- Gautschi: 3.4, 3.7
Link to section 'Module' of 'cutadapt' Module
You can load the modules by:
module load cutadapt
Link to section 'Example job' of 'cutadapt' Example job
To run Cutadapt on our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 1
#SBATCH --job-name=cutadapt
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load cutadapt
cutadapt -a AACCGGTT -o output.fastq input.fastq
eigen
Link to section 'Description' of 'eigen' Description
Eigen is a C++ template library for linear algebra matrices, vectors, numerical solvers, and related algorithms.
Link to section 'Versions' of 'eigen' Versions
- Negishi: 3.3.9
- Anvil: 3.3.9
Link to section 'Module' of 'eigen' Module
You can load the modules by:
module load eigen
emacs
Link to section 'Description' of 'emacs' Description
The Emacs programmable text editor.
Link to section 'Versions' of 'emacs' Versions
- Negishi: 28.2
- Anvil: 27.2
Link to section 'Module' of 'emacs' Module
You can load the modules by:
module load emacs
envi
Link to section 'Description' of 'envi' Description
ENVI is the premier software solution for processing and analyzing geospatial imagery used by scientists, researchers, image analysts, and GIS professionals around the world.
Link to section 'Versions' of 'envi' Versions
- Bell: 5.5.2
- Gilbreth: 5.5.2
Link to section 'Module' of 'envi' Module
You can load the modules by:
module load envi
fastqc
Link to section 'Description' of 'fastqc' Description
A quality control tool for high throughput sequence data.
Link to section 'Versions' of 'fastqc' Versions
- Anvil: 0.11.9, 0.12.1
- Bell: 0.11.9, 0.12.1
- Negishi: 0.11.9, 0.12.1
- Scholar: 0.11.9, 0.12.1
- Gautschi: 0.11.9, 0.12.1
Link to section 'Module' of 'fastqc' Module
You can load the modules by:
module load fastqc
Link to section 'Example job' of 'fastqc' Example job
To run Fastqc on our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 4
#SBATCH --job-name=fastqc
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load fastqc
fastqc -o fastqc_out -t 4 FASTQ1 FASTQ2
fasttree
Link to section 'Description' of 'fasttree' Description
FastTree infers approximately-maximum-likelihood phylogenetic trees from alignments of nucleotide or protein sequences. FastTree can handle alignments with up to a million of sequences in a reasonable amount of time and memory.
Link to section 'Versions' of 'fasttree' Versions
- Anvil: 2.1.10, 2.1.11
- Bell: 2.1.10, 2.1.11
- Negishi: 2.1.10, 2.1.11
- Scholar: 2.1.10, 2.1.11
- Gautschi: 2.1.10, 2.1.11
Link to section 'Module' of 'fasttree' Module
You can load the modules by:
module load fasttree
Link to section 'Example job using single CPU' of 'fasttree' Example job using single CPU
To run FastTree on our our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 20:00:00
#SBATCH -N 1
#SBATCH -n 1
#SBATCH --job-name=fasttree
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load fasttree
FastTree alignmentfile > treefile
Link to section 'Example job using multiple CPUs' of 'fasttree' Example job using multiple CPUs
To run FastTree on our our clusters using multiple CPUs:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 20:00:00
#SBATCH -N 1
#SBATCH -n 24
#SBATCH --job-name=FastTreeMP
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load fasttree
export OMP_NUM_THREADS=24
FastTreeMP alignmentfile > treefile
fastx-toolkit
Link to section 'fastx-toolkit' of 'fastx-toolkit' fastx-toolkit
Link to section 'Description' of 'fastx-toolkit' Description
The FASTX-Toolkit is a collection of command line tools for Short-Reads FASTA/FASTQ files preprocessing.
Link to section 'Versions' of 'fastx-toolkit' Versions
- Anvil: 0.0.14
Link to section 'Commands' of 'fastx-toolkit' Commands
- fasta_clipping_histogram.pl
- fasta_formatter
- fasta_nucleotide_changer
- fastq_masker
- fastq_quality_boxplot_graph.sh
- fastq_quality_converter
- fastq_quality_filter
- fastq_quality_trimmer
- fastq_to_fasta
- fastx_artifacts_filter
- fastx_barcode_splitter.pl
- fastx_clipper
- fastx_collapser
- fastx_nucleotide_distribution_graph.sh
- fastx_nucleotide_distribution_line_graph.sh
- fastx_quality_stats
- fastx_renamer
- fastx_reverse_complement
- fastx_trimmer
- fastx_uncollapser
Link to section 'Module' of 'fastx-toolkit' Module
You can load the modules by:
module load fastx-toolkit
Link to section 'Example job' of 'fastx-toolkit' Example job
To run FASTX-Toolkit on our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 1
#SBATCH --job-name=fastx_toolkit
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load fastx_toolkit
ffmpeg
Link to section 'Description' of 'ffmpeg' Description
FFmpeg is a complete, cross-platform solution to record, convert and stream audio and video.
Link to section 'Versions' of 'ffmpeg' Versions
- Bell: 4.2.2
- Gilbreth: 4.2.1
- Negishi: 4.4.1
- Anvil: 4.2.2
Link to section 'Module' of 'ffmpeg' Module
You can load the modules by:
module load ffmpeg
fftw
Link to section 'Description' of 'fftw' Description
FFTW is a C subroutine library for computing the discrete Fourier transform DFT in one or more dimensions, of arbitrary input size, and of both real and complex data as well as of even/odd data, i.e. the discrete cosine/sine transforms or DCT/DST. We believe that FFTW, which is free software, should become the FFT library of choice for most applications.
Link to section 'Versions' of 'fftw' Versions
- Bell: 3.3.8, 3.3.10
- Scholar: 3.3.10
- Gilbreth: 3.3.7, 3.3.10
- Negishi: 3.3.10
- Anvil: 2.1.5, 3.3.8
- Gautschi: 3.3.10
Link to section 'Module' of 'fftw' Module
You can load the modules by:
module load fftw
gamess
Link to section 'Description' of 'gamess' Description
The General Atomic and Molecular Electronic Structure System (GAMESS) is a general ab initio quantum chemistry package.
Link to section 'Versions' of 'gamess' Versions
- Bell: 18.Aug.2016.R1, 30.Jun.2019.R1
- Scholar: 17.09-r2-libcchem
- Gilbreth: 30.Jun.2019, 17.09-r2-libcchem
Link to section 'Module' of 'gamess' Module
You can load the modules by:
module load gamess
gatk
Link to section 'Description' of 'gatk' Description
Genome Analysis Toolkit Variant Discovery in High-Throughput Sequencing Data
Link to section 'Versions' of 'gatk' Versions
- Anvil: 3.8, 4.1.8.1
- Bell: 3.8
- Negishi: 3.8
- Scholar: 3.8
- Gautschi: 3.8
Link to section 'Commands' of 'gatk' Commands
- gatk
Link to section 'Module' of 'gatk' Module
You can load the modules by:
module load gatk
Link to section 'Example job' of 'gatk' Example job
To run gatk our our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 20:00:00
#SBATCH -N 1
#SBATCH -n 24
#SBATCH -p PartitionName
#SBATCH --job-name=gatk
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load gatk
gatk --java-options "-Xmx12G -XX:ParallelGCThreads=24" HaplotypeCaller -R hg38.fa -I 19P0126636WES.sorted.bam -O 19P0126636WES.HC.vcf --sample-name 19P0126636
gaussian09
Link to section 'Description' of 'gaussian09' Description
Gaussian is a general purpose computational chemistry software package initially released in 1970. It utilizes fundamental laws of quantum mechanics to predict energies, molecular structures, spectroscopic data (NMR, IR, UV) and much more advanced calculations. It provides state-of-the-art capabilities for electronic structure modeling.
Link to section 'Versions' of 'gaussian09' Versions
- Bell: E.01
- Scholar: E.01
Link to section 'Module' of 'gaussian09' Module
You can load the modules by:
module load gaussian09
gaussian16
Link to section 'Description' of 'gaussian16' Description
Gaussian is a general purpose computational chemistry software package initially released in 1970. It utilizes fundamental laws of quantum mechanics to predict energies, molecular structures, spectroscopic data (NMR, IR, UV) and much more advanced calculations. It provides state-of-the-art capabilities for electronic structure modeling.
Link to section 'Versions' of 'gaussian16' Versions
- Bell: B.01
- Scholar: B.01
- Gilbreth: A.03, B.01-gpu
- Negishi: B.01
Link to section 'Module' of 'gaussian16' Module
You can load the modules by:
module load gaussian16
gaussview
Link to section 'Description' of 'gaussview' Description
GaussView is a graphical interface used with Gaussian. It aids in the creation of Gaussian input files, enables the user to run Gaussian calculations from a graphical interface without the need for using a command line instruction, and helps in the interpretation of Gaussian output (e.g., you can use it to plot properties, animate vibrations, visualize computed spectra, etc.).
Link to section 'Versions' of 'gaussview' Versions
- Bell: 5.0.8, 6.0.16
- Scholar: 5.0.8, 6.0.16
- Gilbreth: 6.0.16
- Negishi: 6.0.16
Link to section 'Module' of 'gaussview' Module
You can load the modules by:
module load gaussview
gdal
Link to section 'Description' of 'gdal' Description
GDAL Geospatial Data Abstraction Library is a translator library for raster and vector geospatial data formats that is released under an X/MIT style Open Source license by the Open Source Geospatial Foundation. As a library, it presents a single raster abstract data model and vector abstract data model to the calling application for all supported formats. It also comes with a variety of useful command line utilities for data translation and processing.
Link to section 'Versions' of 'gdal' Versions
- Bell: 2.4.2, 3.4.2, 3.5.3, 3.5.3_sqlite3
- Scholar: 3.9.2-w7ljnyv
- Gilbreth: 2.4.2, 3.5.3, 3.5.3-grib
- Negishi: 2.4.4, 3.5.3
- Anvil: 2.4.4, 3.2.0
Link to section 'Module' of 'gdal' Module
You can load the modules by:
module load gdal
gdb
Link to section 'Description' of 'gdb' Description
GDB, the GNU Project debugger, allows you to see what is going on inside another program while it executes -- or what another program was doing at the moment it crashed.
Link to section 'Versions' of 'gdb' Versions
- Bell: 11.1
- Negishi: 11.1, 12.1
- Anvil: 11.1
Link to section 'Module' of 'gdb' Module
You can load the modules by:
module load gdb
geos
Link to section 'Description' of 'geos' Description
GEOS Geometry Engine - Open Source is a C++ port of the Java Topology Suite JTS. As such, it aims to contain the complete functionality of JTS in C++. This includes all the OpenGIS Simple Features for SQL spatial predicate functions and spatial operators, as well as specific JTS enhanced topology functions.
Link to section 'Versions' of 'geos' Versions
- Bell: 3.8.1, 3.9.4
- Gilbreth: 3.7.2, 3.9.4
- Negishi: 3.9.1
- Anvil: 3.8.1, 3.9.1
Link to section 'Module' of 'geos' Module
You can load the modules by:
module load geos
gmp
Link to section 'Description' of 'gmp' Description
GMP is a free library for arbitrary precision arithmetic, operating on signed integers, rational numbers, and floating-point numbers.
Link to section 'Versions' of 'gmp' Versions
- Bell: 6.1.2, 6.2.1, 6.3.0
- Gilbreth: 6.1.2
- Negishi: 6.2.1
- Anvil: 6.2.1
Link to section 'Module' of 'gmp' Module
You can load the modules by:
module load gmp
gmt
Link to section 'Description' of 'gmt' Description
GMT Generic Mapping Tools is an open source collection of about 80 command-line tools for manipulating geographic and Cartesian data sets including filtering, trend fitting, gridding, projecting, etc. and producing PostScript illustrations ranging from simple x-y plots via contour maps to artificially illuminated surfaces and 3D perspective views.
Link to section 'Versions' of 'gmt' Versions
- Bell: 5.4.4
- Gilbreth: 5.4.4
- Anvil: 6.1.0
- Negishi: 5.4.4, 6.2.0
Link to section 'Module' of 'gmt' Module
You can load the modules by:
module load gmt
gnuplot
Link to section 'Description' of 'gnuplot' Description
Gnuplot is a portable command-line driven graphing utility for Linux, OS/2, MS Windows, OSX, VMS, and many other platforms. The source code is copyrighted but freely distributed i.e., you don't have to pay for it. It was originally created to allow scientists and students to visualize mathematical functions and data interactively, but has grown to support many non-interactive uses such as web scripting. It is also used as a plotting engine by third-party applications like Octave. Gnuplot has been supported and under active development since 1986
Link to section 'Versions' of 'gnuplot' Versions
- Bell: 5.2.8
- Gilbreth: 5.2.7
- Negishi: 5.4.2
- Anvil: 5.4.2
Link to section 'Module' of 'gnuplot' Module
You can load the modules by:
module load gnuplot
gpaw
Link to section 'Description' of 'gpaw' Description
GPAW is a density-functional theory DFT Python code based on the projector-augmented wave PAW method and the atomic simulation environment ASE.
Link to section 'Versions' of 'gpaw' Versions
- Anvil: 21.1.0
Link to section 'Module' of 'gpaw' Module
You can load the modules by:
module load gpaw
grads
Link to section 'Description' of 'grads' Description
The Grid Analysis and Display System (GrADS) is an interactive desktop tool that is used for easy access, manipulation, and visualization of earth science data. GrADS has two data models for handling gridded and station data. GrADS supports many data file formats, including binary (stream or sequential), GRIB (version 1 and 2), NetCDF, HDF (version 4 and 5), and BUFR (for station data).
Link to section 'Versions' of 'grads' Versions
- Bell: 2.2.1
- Gilbreth: 2.2.1
- Negishi: 2.2.1
- Anvil: 2.2.1
Link to section 'Module' of 'grads' Module
You can load the modules by:
module load grads
gromacs
Link to section 'Description' of 'gromacs' Description
GROMACS GROningen MAchine for Chemical Simulations is a molecular dynamics package primarily designed for simulations of proteins, lipids and nucleic acids. It was originally developed in the Biophysical Chemistry department of University of Groningen, and is now maintained by contributors in universities and research centers across the world.
Link to section 'Versions' of 'gromacs' Versions
- Bell: 2018.4, 2019.2, 2020.3, 2022.3
- Scholar: 2018.2, 2020.2, 2021, 2021.3, 2024.3
- Gilbreth: 2018.2, 2018.4, 2019.2, 2020.2, 2021, 2021.3, 2024.1
- Negishi: 2020.3, 2022.3
- Anvil: 2019.6, 2020.2, 2021, 2021.2, 2021.3, 2024.1
- Gautschi: 2024.3
Link to section 'Module' of 'gromacs' Module
You can check available gromacs version by:
module spider gromacs
You can check how to load the gromacs module by the module's full name:
module spider gromacs/XXXX
Note: RCAC also installed some containerized gromacs modules.
To use these containerized modules, please following the instructions in the output of "module spider gromacs/XXXX"
You can load the modules by:
module load gromacs # for default version
module load gromacs/XXXX # for specific version
Link to section 'Usage' of 'gromacs' Usage
The GROMACS executable is gmx_mpi and you can use gmx help commands for help on a command.
For more details about how to run GROMACS, please check GROMACS.
Link to section 'Example job' of 'gromacs' Example job
#!/bin/bash
# FILENAME: myjobsubmissionfile
#SBATCH --nodes=2 # Total # of nodes
#SBATCH --ntasks=256 # Total # of MPI tasks
#SBATCH --time=1:30:00 # Total run time limit (hh:mm:ss)
#SBATCH -J myjobname # Job name
#SBATCH -o myjob.o%j # Name of stdout output file
#SBATCH -e myjob.e%j # Name of stderr error file
# Manage processing environment, load compilers and applications.
module purge
module load gcc/XXXX openmpi/XXXX # or module load intel/XXXX impi/XXXX | depends on the output of "module spider gromacs/XXXX"
module load gromacs/XXXX
module list
# Launch MPI code
gmx_mpi pdb2gmx -f my.pdb -o my_processed.gro -water spce
gmx_mpi grompp -f my.mdp -c my_processed.gro -p topol.top -o topol.tpr
srun -n $SLURM_NTASKS gmx_mpi mdrun -s topol.tpr
Link to section 'Note' of 'gromacs' Note
Using mpirun -np $SLURM_NTASKS gmx_mpi or mpiexex -np $SLURM_NTASKS gmx_mpi may not work for non-exclusive jobs on some clusters. Use srun -n $SLURM_NTASKS gmx_mpi or mpirun gmx_mpi instead. mpirun gmx_mpi without specifying the number of ranks will automatically pick up the number of SLURM_NTASKS and works fine.
gsl
Link to section 'Description' of 'gsl' Description
The GNU Scientific Library GSL is a numerical library for C and C++ programmers. It is free software under the GNU General Public License. The library provides a wide range of mathematical routines such as random number generators, special functions and least-squares fitting. There are over 1000 functions in total with an extensive test suite.
Link to section 'Versions' of 'gsl' Versions
- Bell: 2.4
- Gilbreth: 2.4
- Negishi: 2.4, 2.7.1
- Anvil: 2.4
Link to section 'Module' of 'gsl' Module
You can load the modules by:
module load gsl
gurobi
Link to section 'Description' of 'gurobi' Description
The Gurobi Optimizer was designed from the ground up to be the fastest, most powerful solver available for your LP, QP, QCP, and MIP MILP, MIQP, and MIQCP problems.
Link to section 'Versions' of 'gurobi' Versions
- Bell: 9.0.1, 9.5.1, 10.0.1
- Scholar: 11.0.3
- Anvil: 9.5.1
- Negishi: 10.0.1
- Gautschi: 11.0.3
Link to section 'Module' of 'gurobi' Module
You can load the modules by:
module load gurobi
gym
Link to section 'Description' of 'gym' Description
The OpenAI Gym: A toolkit for developing and comparing your reinforcement learning agents.
Link to section 'Versions' of 'gym' Versions
- Bell: 0.17.3
- Gilbreth: 0.10.9, 0.18.0
Link to section 'Module' of 'gym' Module
You can load the modules by:
module load learning
module load gym
hadoop
Link to section 'Description' of 'hadoop' Description
The Apache Hadoop software library is a framework that allows for the distributed processing of large data sets across clusters of computers using simple programming models.
Link to section 'Versions' of 'hadoop' Versions
- Bell: 2.7.7
- Gilbreth: 2.7.7
- Negishi: 3.3.2
- Anvil: 3.3.0
Link to section 'Module' of 'hadoop' Module
You can load the modules by:
module load hadoop
hdf
Link to section 'Description' of 'hdf' Description
HDF4 also known as HDF is a library and multi-object file format for storing and managing data between machines.
Link to section 'Versions' of 'hdf' Versions
- Bell: 4.2.15
- Gilbreth: 4.2.14
- Negishi: 4.2.15
- Anvil: 4.2.15
Link to section 'Module' of 'hdf' Module
You can load the modules by:
module load hdf
hdf5
Link to section 'Description' of 'hdf5' Description
HDF5 is a data model, library, and file format for storing and managing data. It supports an unlimited variety of datatypes, and is designed for flexible and efficient I/O and for high volume and complex data.
Link to section 'Versions' of 'hdf5' Versions
- Bell: 1.8.21, 1.10.6, 1.13.2
- Scholar: 1.14.3
- Gilbreth: 1.8.16, 1.10.5, 1.14.1
- Negishi: 1.13.2
- Anvil: 1.10.7
- Gautschi: 1.14.3
Link to section 'Module' of 'hdf5' Module
You can load the modules by:
module load hdf5
hpctoolkit
Link to section 'Description' of 'hpctoolkit' Description
HPCToolkit is an integrated suite of tools for measurement and analysis of program performance on computers ranging from multicore desktop systems to the nations largest supercomputers. By using statistical sampling of timers and hardware performance counters, HPCToolkit collects accurate measurements of a programs work, resource consumption, and inefficiency and attributes them to the full calling context in which they occur.
Link to section 'Versions' of 'hpctoolkit' Versions
- Negishi: 2022.05.15
- Anvil: 2021.03.01
Link to section 'Module' of 'hpctoolkit' Module
You can load the modules by:
module load hpctoolkit
hspice
Link to section 'Description' of 'hspice' Description
Hspice is a device level circuit simulator. Hspice takes a spice file as input and produces output describing the requested simulation of the circuit. It can also produce output files to be used by the AWAVES post processor.
Link to section 'Versions' of 'hspice' Versions
- Bell: 2017.12, 2019.06, 2020.12
- Negishi: 2020.12
Link to section 'Module' of 'hspice' Module
You can load the modules by:
module load hspice
htseq
Link to section 'Description' of 'htseq' Description
HTSeq is a Python package that provides infrastructure to process data from high-throughput sequencing assays.
Link to section 'Versions' of 'htseq' Versions
- Anvil: 0.11.2, 0.13.5, 1.99.2, 2.0.1, 2.0.2
- Bell: 0.13.5, 1.99.2, 2.0.1, 2.0.2
- Negishi: 0.13.5, 1.99.2, 2.0.1, 2.0.2
- Scholar: 0.13.5, 1.99.2, 2.0.1, 2.0.2, 2.0.2-py310
- Gautschi: 0.13.5, 1.99.2, 2.0.1, 2.0.2, 2.0.2-py310
Link to section 'Commands' of 'htseq' Commands
- htseq-count
- htseq-qa
Link to section 'Module' of 'htseq' Module
You can load the modules by:
module load htseq
Link to section 'Example job' of 'htseq' Example job
To run HTSeq on our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 1
#SBATCH --job-name=htseq
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load htseq
htseq-count input.bam ref.gtf > test.out
hwloc
Link to section 'Description' of 'hwloc' Description
The Hardware Locality hwloc software project.
Link to section 'Versions' of 'hwloc' Versions
- Anvil: 1.11.13
Link to section 'Module' of 'hwloc' Module
You can load the modules by:
module load hwloc
hyper-shell
Link to section 'Description' of 'hyper-shell' Description
Process shell commands over a distributed, asynchronous queue.
Documentation: https://hyper-shell.readthedocs.io/en/latest/
Link to section 'Versions' of 'hyper-shell' Versions
- Bell: 1.8.3, 2.0.2, 2.4.0, 2.5.1
- Scholar: 2.5.2
- Gilbreth: 1.8.3, 2.0.2, 2.4.0, 2.5.1
- Negishi: 2.0.2, 2.1.0, 2.4.0, 2.5.1
- Anvil: 2.0.2, 2.2.0, 2.4.0, 2.5.1
- Gautschi: 2.5.2
Link to section 'Module' of 'hyper-shell' Module
You can load the modules by:
module load hyper-shell
hypre
Link to section 'Description' of 'hypre' Description
Hypre is a library of high performance preconditioners that features parallel multigrid methods for both structured and unstructured grid problems.
Link to section 'Versions' of 'hypre' Versions
- Bell: 2.18.1
Link to section 'Module' of 'hypre' Module
You can load the modules by:
module load hypre
idl
Link to section 'Description' of 'idl' Description
IDL is a data analysis language that provides powerful, core visualization and analysis functionality, and capabilities that allow data analysts and developers to leverage IDL's power in multiple software environments.
Link to section 'Versions' of 'idl' Versions
- Bell: 8.7
- Gilbreth: 8.7
Link to section 'Module' of 'idl' Module
You can load the modules by:
module load idl
intel-mkl
Link to section 'Description' of 'intel-mkl' Description
Intel Math Kernel Library (MKL) contains ScaLAPACK, LAPACK, Sparse Solver, BLAS, Sparse BLAS, CBLAS, GMP, FFTs, DFTs, VSL, VML, and Interval Arithmetic routines. MKL resides in the directory stored in the environment variable MKL_HOME, after loading a version of the Intel compiler with module.
Link to section 'Versions' of 'intel-mkl' Versions
- Bell: 2017.1.132, 2019.5.281, 2024.1
- Negishi: 2019.9.304
- Anvil: 2019.5.281, 2020.4.304
Link to section 'Module' of 'intel-mkl' Module
You can load the modules by:
module load intel-mkl
By using module load to load an Intel compiler your environment will have several variables set up to help link applications with MKL. Here are some example combinations of simplified linking options:
$ module load intel
$ echo $LINK_LAPACK
-L${MKL_HOME}/lib/intel64 -lmkl_intel_lp64 -lmkl_intel_thread -lmkl_core -liomp5 -lpthread
$ echo $LINK_LAPACK95
-L${MKL_HOME}/lib/intel64 -lmkl_lapack95_lp64 -lmkl_blas95_lp64 -lmkl_intel_lp64 -lmkl_intel_thread -lmkl_core -liomp5 -lpthread
RCAC recommends you use the provided variables to define MKL linking options in your compiling procedures. The Intel compiler modules also provide two other environment variables, LINK_LAPACK_STATIC and LINK_LAPACK95_STATIC that you may use if you need to link MKL statically.
RCAC recommends that you use dynamic linking of libguide. If so, define LD_LIBRARY_PATH such that you are using the correct version of libguide at run time. If you use static linking of libguide, then:
- If you use the Intel compilers, link in the libguide version that comes with the compiler (use the -openmp option).
- If you do not use the Intel compilers, link in the libguide version that comes with the Intel MKL above.
Here are some more documentation from other sources on the Intel MKL:
intel-oneapi-mkl
Link to section 'Description' of 'intel-oneapi-mkl' Description
Intel oneAPI Math Kernel Library Intel oneMKL; formerly Intel Math Kernel Library or Intel MKL, is a library of optimized math routines for science, engineering, and financial applications. Core math functions include BLAS, LAPACK, ScaLAPACK, sparse solvers, fast Fourier transforms, and vector math.
Link to section 'Versions' of 'intel-oneapi-mkl' Versions
- Negishi: 2023.0.0
Link to section 'Module' of 'intel-oneapi-mkl' Module
You can load the modules by:
module load intel-oneapi-mkl
intel-oneapi-tbb
Link to section 'Description' of 'intel-oneapi-tbb' Description
Intel oneAPI Threading Building Blocks oneTBB is a flexible performance library that simplifies the work of adding parallelism to complex applications across accelerated architectures, even if you are not a threading expert.
Link to section 'Versions' of 'intel-oneapi-tbb' Versions
- Negishi: 2021.8.0
Link to section 'Module' of 'intel-oneapi-tbb' Module
You can load the modules by:
module load intel-oneapi-tbb
julia
Link to section 'Description' of 'julia' Description
Julia is a flexible dynamic language, appropriate for scientific and numerical computing, with performance comparable to traditional statically-typed languages. One can write code in Julia that is nearly as fast as C. Julia features optional typing, multiple dispatch, and good performance, achieved using type inference and just-in-time (JIT) compilation, implemented using LLVM. It is multi-paradigm, combining features of imperative, functional, and object-oriented programming.
Link to section 'Versions' of 'julia' Versions
- Bell: 1.7.1, 1.8.1, 1.9.3
- Gilbreth: 1.7.1, 1.8.5, 1.9.3
- Negishi: 1.8.5, 1.9.3
- Anvil: 1.6.2, 1.9.3
- Gautschi: 1.11.1
- Scholar: 1.5.0, 2.4.2
Link to section 'Module' of 'julia' Module
You can load the modules by:
module load julia
Link to section 'Package installation' of 'julia' Package installation
Users do not have write permission to the default julia package installation destination. However, users can install packages into home directory under ~/.julia.
Users can side step this by explicitly defining where to put julia packages:
$ export JULIA_DEPOT_PATH=$HOME/.julia
$ julia -e 'using Pkg; Pkg.add("PackageName")'
jupyter
Link to section 'Description' of 'jupyter' Description
Complete Jupyter Hub/Lab/Notebook environment.
Link to section 'Versions' of 'jupyter' Versions
- Anvil: 2.0.0
- Negishi: 3.1.1
- Gautschi: 1.1.1
Link to section 'Module' of 'jupyter' Module
You can load the modules by:
module load jupyter
jupyterhub
Link to section 'Description' of 'jupyterhub' Description
Complete Jupyter Hub/Lab/Notebook environment.
Link to section 'Versions' of 'jupyterhub' Versions
- Bell: 2.0.0
- Scholar: 2.0.0
- Gilbreth: 2.0.0
Link to section 'Module' of 'jupyterhub' Module
You can load the modules by:
module load jupyterhub
lammps
Link to section 'Description' of 'lammps' Description
LAMMPS is a classical molecular dynamics code with a focus on materials modelling. It’s an acronym for Large-scale Atomic/Molecular Massively Parallel Simulator.
LAMMPS has potentials for solid-state materials (metals, semiconductors) and soft matter (biomolecules, polymers) and coarse-grained or mesoscopic systems. It can be used to model atoms or, more generically, as a parallel particle simulator at the atomic, meso, or continuum scale.
Link to section 'Versions' of 'lammps' Versions
- Bell: 20200721, 20201029, 2022.5.04
- Scholar: 10Feb2021, 15Jun2020, 24Oct2018, 29Oct2020, 20240829
- Gilbreth: 10Feb2021, 15Jun2020, 24Oct2018, 29Oct2020
- Negishi: 2022.5.04, 20220623
- Anvil: 20210310, 20230802
- Gautschi: 20240829
Link to section 'Module' of 'lammps' Module
You can check available lammps version by:
module spider lammps
You can check how to load the lammps module by the module's full name:
module spider lammps/XXXX
You can load the modules by:
module load lammps # for default version
module load lammps/XXXX # for specific version
Link to section 'Usage' of 'lammps' Usage
LAMMPS reads command lines from an input file like "in.file". The LAMMPS executable is lmp, to run the lammps input file, use the -in command:
lmp -in in.file
For more details about how to run LAMMPS, please check LAMMPS.
Link to section 'Example job' of 'lammps' Example job
#!/bin/bash
# FILENAME: myjobsubmissionfile
#SBATCH --nodes=2 # Total # of nodes
#SBATCH --ntasks=256 # Total # of MPI tasks
#SBATCH --time=1:30:00 # Total run time limit (hh:mm:ss)
#SBATCH -J myjobname # Job name
#SBATCH -o myjob.o%j # Name of stdout output file
#SBATCH -e myjob.e%j # Name of stderr error file
# Manage processing environment, load compilers and applications.
module purge
module load gcc/XXXX openmpi/XXXX # or module load intel/XXXX impi/XXXX | depends on the output of "module spider lammps/XXXX"
module load lammps/XXXX
module list
# Launch MPI code
srun -n $SLURM_NTASKS lmp
Link to section 'Note' of 'lammps' Note
Using mpirun -np $SLURM_NTASKS lmp or mpiexex -np $SLURM_NTASKS lmp may not work for non-exclusive jobs on some clusters. Use srun -n $SLURM_NTASKS lmp or mpirun lmp instead. mpirun lmp without specifying the number of ranks will automatically pick up the number of SLURM_NTASKS and works fine.
launcher
Link to section 'Description' of 'launcher' Description
Framework for running large collections of serial or multi-threaded applications
Link to section 'Versions' of 'launcher' Versions
- Anvil: 3.9
- Negishi: 3.9
- Gautschi: 3.9
Link to section 'Module' of 'launcher' Module
You can load the modules by:
module load launcher
learning
Link to section 'Description' of 'learning' Description
The learning module loads the prerequisites (such as anaconda and cudnn ) and makes ML applications visible to the user
Link to section 'Versions' of 'learning' Versions
- Bell: conda-2020.11-py38-cpu
- Gilbreth: conda-5.1.0-py27-cpu, conda-5.1.0-py27-gpu, conda-5.1.0-py36-cpu, conda-5.1.0-py36-gpu, conda-2020.11-py38-cpu, conda-2020.11-py38-gpu
- Anvil: conda-2021.05-py38-gpu
Link to section 'Module' of 'learning' Module
You can load the modules by:
module load learning
Link to section 'Example job' of 'learning' Example job
This is the example jobscript for our cluster `Gilbreth`:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 20:00:00
#SBATCH -N 1
#SBATCH -n 10
#SBATCH --gpus-per-node=1
#SBATCH -p PartitionName
#SBATCH --job-name=learning
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module --force purge
module load learning/conda-2020.11-py38-gpu
module load ml-toolkit-gpu/pytorch/1.7.1
python torch.py
libfabric
Link to section 'Description' of 'libfabric' Description
The Open Fabrics Interfaces OFI is a framework focused on exporting fabric communication services to applications.
Link to section 'Versions' of 'libfabric' Versions
- Negishi: 1.12.0
- Anvil: 1.12.0
Link to section 'Module' of 'libfabric' Module
You can load the modules by:
module load libfabric
libflame
Link to section 'Description' of 'libflame' Description
libflame is a portable library for dense matrix computations, providing much of the functionality present in LAPACK, developed by current and former members of the Science of High-Performance Computing SHPC group in the Institute for Computational Engineering and Sciences at The University of Texas at Austin. libflame includes a compatibility layer, lapack2flame, which includes a complete LAPACK implementation.
Link to section 'Versions' of 'libflame' Versions
- Negishi: 5.2.0
- Anvil: 5.2.0
- Gautschi: 5.2.0
- Scholar: 5.2.0
Link to section 'Module' of 'libflame' Module
You can load the modules by:
module load libflame
libiconv
Link to section 'Description' of 'libiconv' Description
GNU libiconv provides an implementation of the iconv function and the iconv program for character set conversion.
Link to section 'Versions' of 'libiconv' Versions
- Bell: 1.16, 1.17
- Gilbreth: 1.16
- Negishi: 1.16
- Anvil: 1.16
Link to section 'Module' of 'libiconv' Module
You can load the modules by:
module load libiconv
libmesh
Link to section 'Description' of 'libmesh' Description
The libMesh library provides a framework for the numerical simulation of partial differential equations using arbitrary unstructured discretizations on serial and parallel platforms.
Link to section 'Versions' of 'libmesh' Versions
- Negishi: 1.7.1
- Anvil: 1.6.2
Link to section 'Module' of 'libmesh' Module
You can load the modules by:
module load libmesh
libszip
Link to section 'Description' of 'libszip' Description
Szip is an implementation of the extended-Rice lossless compression algorithm.
Link to section 'Versions' of 'libszip' Versions
- Bell: 2.1.1
- Negishi: 2.1.1
- Anvil: 2.1.1
Link to section 'Module' of 'libszip' Module
You can load the modules by:
module load libszip
libtiff
Link to section 'Description' of 'libtiff' Description
LibTIFF - Tag Image File Format TIFF Library and Utilities.
Link to section 'Versions' of 'libtiff' Versions
- Bell: 4.0.10, 4.4.0, 4.6.0
- Gilbreth: 4.0.10
- Negishi: 4.4.0
- Anvil: 4.1.0
Link to section 'Module' of 'libtiff' Module
You can load the modules by:
module load libtiff
libx11
Link to section 'Description' of 'libx11' Description
Xlib − C Language X Interface is a reference guide to the low-level C language interface to the X Window System protocol. It is neither a tutorial nor a user’s guide to programming the X Window System. Rather, it provides a detailed description of each function in the library as well as a discussion of the related background information.
Link to section 'Versions' of 'libx11' Versions
- Anvil: 1.7.0
- Negishi: 1.7.0
Link to section 'Module' of 'libx11' Module
You can load the modules by:
module load libx11
libxml2
Link to section 'Description' of 'libxml2' Description
Libxml2 is the XML C parser and toolkit developed for the Gnome project but usable outside of the Gnome platform, it is free software available under the MIT License.
Link to section 'Versions' of 'libxml2' Versions
- Bell: 2.9.9, 2.10.3
- Gilbreth: 2.9.9
- Negishi: 2.10.1
- Anvil: 2.9.10
Link to section 'Module' of 'libxml2' Module
You can load the modules by:
module load libxml2
mathematica
Link to section 'Description' of 'mathematica' Description
Mathematica is a technical computing environment and programming language with strong symbolic and numerical abilities.
Link to section 'Versions' of 'mathematica' Versions
- Bell: 11.3, 12.1, 12.3, 13.1, 14.1
- Scholar: 14.1
- Gilbreth: 11.3, 12.1, 12.3, 13.1, 14.1
- Negishi: 13.1, 14.1
- Gautschi: 14.1
Link to section 'Module' of 'mathematica' Module
You can load the modules by:
module load mathematica
Link to section 'Running Mathematica' of 'mathematica' Running Mathematica
Users can run Mathematica GUI in interactive jobs or run it as batch jobs.Link to section 'Interactive jobs' of 'mathematica' Interactive jobs
sinteractive -N1 -n24 -t4:00:00 -A standby
module load mathematica
Mathematica
Notice: The executable name has changed from Mathematica to WolframNB in version 14.1
Link to section 'Batch job' of 'mathematica' Batch job
To submit a sbatch job on our clusters:
#!/bin/bash
#SBATCH -A XXX
#SBATCH -t 10:00:00
#SBATCH -N 1
#SBATCH -n 24
#SBATCH --job-name=mathematica
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load mathematica
math -noprompt < input.m
matlab
Link to section 'Description' of 'matlab' Description
MATLAB MATrix LABoratory is a multi-paradigm numerical computing environment and fourth-generation programming language. A proprietary programming language developed by MathWorks, MATLAB allows matrix manipulations, plotting of functions and data, implementation of algorithms, creation of user interfaces, and interfacing with programs written in other languages, including C, C++, C#, Java, Fortran and Python.
Link to section 'Versions' of 'matlab' Versions
- Bell: R2019a, R2020a, R2020b, R2021b, R2022a, R2023a
- Scholar: R2024a
- Gilbreth: R2017a, R2018a, R2019a, R2020a, R2022a, R2023a
- Negishi: R2021b, R2022a, R2023a
- Anvil: R2020b, R2021b, R2022a, R2023a
- Gautschi: R2024a
Link to section 'Module' of 'matlab' Module
You can load the modules by:
module load matlab
MATLAB, Simulink, Compiler, and several of the optional toolboxes are available to faculty, staff, and students. To see the kind and quantity of all MATLAB licenses plus the number that you are currently using you can use the matlab_licenses command:
$ module load matlab
$ matlab_licenses
The MATLAB client can be run in the front-end for application development, however, computationally intensive jobs must be run on compute nodes.
The following sections provide several examples illustrating how to submit MATLAB jobs to a Linux compute cluster.
Matlab Script (.m File)
This section illustrates how to submit a small, serial, MATLAB program as a job to a batch queue. This MATLAB program prints the name of the run host and gets three random numbers.
Prepare a MATLAB script myscript.m, and a MATLAB function file myfunction.m:
% FILENAME: myscript.m
% Display name of compute node which ran this job.
[c name] = system('hostname');
fprintf('\n\nhostname:%s\n', name);
% Display three random numbers.
A = rand(1,3);
fprintf('%f %f %f\n', A);
quit;
% FILENAME: myfunction.m
function result = myfunction ()
% Return name of compute node which ran this job.
[c name] = system('hostname');
result = sprintf('hostname:%s', name);
% Return three random numbers.
A = rand(1,3);
r = sprintf('%f %f %f', A);
result=strvcat(result,r);
end
Also, prepare a job submission file, here named myjob.sub. Run with the name of the script:
#!/bin/bash
# FILENAME: myjob.sub
echo "myjob.sub"
# Load module, and set up environment for Matlab to run
module load matlab
unset DISPLAY
# -nodisplay: run MATLAB in text mode; X11 server not needed
# -singleCompThread: turn off implicit parallelism
# -r: read MATLAB program; use MATLAB JIT Accelerator
# Run Matlab, with the above options and specifying our .m file
matlab -nodisplay -singleCompThread -r myscript
myjob.sub
< M A T L A B (R) >
Copyright 1984-2011 The MathWorks, Inc.
R2011b (7.13.0.564) 64-bit (glnxa64)
August 13, 2011
To get started, type one of these: helpwin, helpdesk, or demo.
For product information, visit www.mathworks.com.
hostname:resource-a001.rcac.purdue.edu
0.814724 0.905792 0.126987
Output shows that a processor core on one compute node (resource-a001) processed the job. Output also displays the three random numbers.
For more information about MATLAB:
Implicit Parallelism
MATLAB implements implicit parallelism which is automatic multithreading of many computations, such as matrix multiplication, linear algebra, and performing the same operation on a set of numbers. This is different from the explicit parallelism of the Parallel Computing Toolbox.
MATLAB offers implicit parallelism in the form of thread-parallel enabled functions. Since these processor cores, or threads, share a common memory, many MATLAB functions contain multithreading potential. Vector operations, the particular application or algorithm, and the amount of computation (array size) contribute to the determination of whether a function runs serially or with multithreading.
When your job triggers implicit parallelism, it attempts to allocate its threads on all processor cores of the compute node on which the MATLAB client is running, including processor cores running other jobs. This competition can degrade the performance of all jobs running on the node.
When you know that you are coding a serial job but are unsure whether you are using thread-parallel enabled operations, run MATLAB with implicit parallelism turned off. Beginning with the R2009b, you can turn multithreading off by starting MATLAB with -singleCompThread:
$ matlab -nodisplay -singleCompThread -r mymatlabprogram
When you are using implicit parallelism, make sure you request exclusive access to a compute node, as MATLAB has no facility for sharing nodes.
For more information about MATLAB's implicit parallelism:
Profile Manager
MATLAB offers two kinds of profiles for parallel execution: the 'local' profile and user-defined cluster profiles. The 'local' profile runs a MATLAB job on the processor core(s) of the same compute node, or front-end, that is running the client. To run a MATLAB job on compute node(s) different from the node running the client, you must define a Cluster Profile using the Cluster Profile Manager.
To prepare a user-defined cluster profile, use the Cluster Profile Manager in the Parallel menu. This profile contains the scheduler details (queue, nodes, processors, walltime, etc.) of your job submission. Ultimately, your cluster profile will be an argument to MATLAB functions like batch().
For your convenience, a generic cluster profile is provided that can be downloaded: myslurmprofile.settings
Please note that modifications are very likely to be required to make myslurmprofile.settings work. You may need to change values for number of nodes, number of workers, walltime, and submission queue specified in the file. As well, the generic profile itself depends on the particular job scheduler on the cluster, so you may need to download or create two or more generic profiles under different names. Each time you run a job using a Cluster Profile, make sure the specific profile you are using is appropriate for the job and the cluster.
To import the profile, start a MATLAB session and select Manage Cluster Profiles... from the Parallel menu. In the Cluster Profile Manager, select Import, navigate to the folder containing the profile, select myslurmprofile.settings and click OK. Remember that the profile will need to be customized for your specific needs. If you have any questions, please contact us.
For detailed information about MATLAB's Parallel Computing Toolbox, examples, demos, and tutorials:
Parallel Computing Toolbox (parfor)
The MATLAB Parallel Computing Toolbox (PCT) extends the MATLAB language with high-level, parallel-processing features such as parallel for loops, parallel regions, message passing, distributed arrays, and parallel numerical methods. It offers a shared-memory computing environment running on the local cluster profile in addition to your MATLAB client. Moreover, the MATLAB Distributed Computing Server (DCS) scales PCT applications up to the limit of your DCS licenses.
This section illustrates the fine-grained parallelism of a parallel for loop (parfor) in a pool job.
The following examples illustrate a method for submitting a small, parallel, MATLAB program with a parallel loop (parfor statement) as a job to a queue. This MATLAB program prints the name of the run host and shows the values of variables numlabs and labindex for each iteration of the parfor loop.
This method uses the job submission command to submit a MATLAB client which calls the MATLAB batch() function with a user-defined cluster profile.
Prepare a MATLAB pool program in a MATLAB script with an appropriate filename, here named myscript.m:
% FILENAME: myscript.m
% SERIAL REGION
[c name] = system('hostname');
fprintf('SERIAL REGION: hostname:%s\n', name)
numlabs = parpool('poolsize');
fprintf(' hostname numlabs labindex iteration\n')
fprintf(' ------------------------------- ------- -------- ---------\n')
tic;
% PARALLEL LOOP
parfor i = 1:8
[c name] = system('hostname');
name = name(1:length(name)-1);
fprintf('PARALLEL LOOP: %-31s %7d %8d %9d\n', name,numlabs,labindex,i)
pause(2);
end
% SERIAL REGION
elapsed_time = toc; % get elapsed time in parallel loop
fprintf('\n')
[c name] = system('hostname');
name = name(1:length(name)-1);
fprintf('SERIAL REGION: hostname:%s\n', name)
fprintf('Elapsed time in parallel loop: %f\n', elapsed_time)
The execution of a pool job starts with a worker executing the statements of the first serial region up to the parfor block, when it pauses. A set of workers (the pool) executes the parfor block. When they finish, the first worker resumes by executing the second serial region. The code displays the names of the compute nodes running the batch session and the worker pool.
Prepare a MATLAB script that calls MATLAB function batch() which makes a four-lab pool on which to run the MATLAB code in the file myscript.m. Use an appropriate filename, here named mylclbatch.m:
% FILENAME: mylclbatch.m
!echo "mylclbatch.m"
!hostname
pjob=batch('myscript','Profile','myslurmprofile','Pool',4,'CaptureDiary',true);
wait(pjob);
diary(pjob);
quit;
Prepare a job submission file with an appropriate filename, here named myjob.sub:
#!/bin/bash
# FILENAME: myjob.sub
echo "myjob.sub"
hostname
module load matlab
unset DISPLAY
matlab -nodisplay -r mylclbatch
One processor core runs myjob.sub and mylclbatch.m.
Once this job starts, a second job submission is made.
myjob.sub
< M A T L A B (R) >
Copyright 1984-2013 The MathWorks, Inc.
R2013a (8.1.0.604) 64-bit (glnxa64)
February 15, 2013
To get started, type one of these: helpwin, helpdesk, or demo.
For product information, visit www.mathworks.com.
mylclbatch.mresource-a000.rcac.purdue.edu
SERIAL REGION: hostname:resource-a000.rcac.purdue.edu
hostname numlabs labindex iteration
------------------------------- ------- -------- ---------
PARALLEL LOOP: resource-a001.rcac.purdue.edu 4 1 2
PARALLEL LOOP: resource-a002.rcac.purdue.edu 4 1 4
PARALLEL LOOP: resource-a001.rcac.purdue.edu 4 1 5
PARALLEL LOOP: resource-a002.rcac.purdue.edu 4 1 6
PARALLEL LOOP: resource-a003.rcac.purdue.edu 4 1 1
PARALLEL LOOP: resource-a003.rcac.purdue.edu 4 1 3
PARALLEL LOOP: resource-a004.rcac.purdue.edu 4 1 7
PARALLEL LOOP: resource-a004.rcac.purdue.edu 4 1 8
SERIAL REGION: hostname:resource-a001.rcac.purdue.edu
Elapsed time in parallel loop: 5.411486
To scale up this method to handle a real application, increase the wall time in the submission command to accommodate a longer running job. Secondly, increase the wall time of myslurmprofile by using the Cluster Profile Manager in the Parallel menu to enter a new wall time in the property SubmitArguments.
For more information about MATLAB Parallel Computing Toolbox:
Parallel Toolbox (spmd)
The MATLAB Parallel Computing Toolbox (PCT) extends the MATLAB language with high-level, parallel-processing features such as parallel for loops, parallel regions, message passing, distributed arrays, and parallel numerical methods. It offers a shared-memory computing environment with a maximum of eight MATLAB workers (labs, threads; versions R2009a) and 12 workers (labs, threads; version R2011a) running on the local configuration in addition to your MATLAB client. Moreover, the MATLAB Distributed Computing Server (DCS) scales PCT applications up to the limit of your DCS licenses.
This section illustrates how to submit a small, parallel, MATLAB program with a parallel region (spmd statement) as a MATLAB pool job to a batch queue.
This example uses the submission command to submit to compute nodes a MATLAB client which interprets a Matlab .m with a user-defined cluster profile which scatters the MATLAB workers onto different compute nodes. This method uses the MATLAB interpreter, the Parallel Computing Toolbox, and the Distributed Computing Server; so, it requires and checks out six licenses: one MATLAB license for the client running on the compute node, one PCT license, and four DCS licenses. Four DCS licenses run the four copies of the spmd statement. This job is completely off the front end.
Prepare a MATLAB script called myscript.m:
% FILENAME: myscript.m
% SERIAL REGION
[c name] = system('hostname');
fprintf('SERIAL REGION: hostname:%s\n', name)
p = parpool('4');
fprintf(' hostname numlabs labindex\n')
fprintf(' ------------------------------- ------- --------\n')
tic;
% PARALLEL REGION
spmd
[c name] = system('hostname');
name = name(1:length(name)-1);
fprintf('PARALLEL REGION: %-31s %7d %8d\n', name,numlabs,labindex)
pause(2);
end
% SERIAL REGION
elapsed_time = toc; % get elapsed time in parallel region
delete(p);
fprintf('\n')
[c name] = system('hostname');
name = name(1:length(name)-1);
fprintf('SERIAL REGION: hostname:%s\n', name)
fprintf('Elapsed time in parallel region: %f\n', elapsed_time)
quit;
Prepare a job submission file with an appropriate filename, here named myjob.sub. Run with the name of the script:
#!/bin/bash
# FILENAME: myjob.sub
echo "myjob.sub"
module load matlab
unset DISPLAY
matlab -nodisplay -r myscript
Run MATLAB to set the default parallel configuration to your job configuration:
$ matlab -nodisplay
>> parallel.defaultClusterProfile('myslurmprofile');
>> quit;
$
Once this job starts, a second job submission is made.
myjob.sub
< M A T L A B (R) >
Copyright 1984-2011 The MathWorks, Inc.
R2011b (7.13.0.564) 64-bit (glnxa64)
August 13, 2011
To get started, type one of these: helpwin, helpdesk, or demo.
For product information, visit www.mathworks.com.
SERIAL REGION: hostname:resource-a001.rcac.purdue.edu
Starting matlabpool using the 'myslurmprofile' profile ... connected to 4 labs.
hostname numlabs labindex
------------------------------- ------- --------
Lab 2:
PARALLEL REGION: resource-a002.rcac.purdue.edu 4 2
Lab 1:
PARALLEL REGION: resource-a001.rcac.purdue.edu 4 1
Lab 3:
PARALLEL REGION: resource-a003.rcac.purdue.edu 4 3
Lab 4:
PARALLEL REGION: resource-a004.rcac.purdue.edu 4 4
Sending a stop signal to all the labs ... stopped.
SERIAL REGION: hostname:resource-a001.rcac.purdue.edu
Elapsed time in parallel region: 3.382151
Output shows the name of one compute node (a001) that processed the job submission file myjob.sub and the two serial regions. The job submission scattered four processor cores (four MATLAB labs) among four different compute nodes (a001,a002,a003,a004) that processed the four parallel regions. The total elapsed time demonstrates that the jobs ran in parallel.
For more information about MATLAB Parallel Computing Toolbox:
Distributed Computing Server (parallel job)
The MATLAB Parallel Computing Toolbox (PCT) enables a parallel job via the MATLAB Distributed Computing Server (DCS). The tasks of a parallel job are identical, run simultaneously on several MATLAB workers (labs), and communicate with each other. This section illustrates an MPI-like program.
This section illustrates how to submit a small, MATLAB parallel job with four workers running one MPI-like task to a batch queue. The MATLAB program broadcasts an integer to four workers and gathers the names of the compute nodes running the workers and the lab IDs of the workers.
This example uses the job submission command to submit a Matlab script with a user-defined cluster profile which scatters the MATLAB workers onto different compute nodes. This method uses the MATLAB interpreter, the Parallel Computing Toolbox, and the Distributed Computing Server; so, it requires and checks out six licenses: one MATLAB license for the client running on the compute node, one PCT license, and four DCS licenses. Four DCS licenses run the four copies of the parallel job. This job is completely off the front end.
Prepare a MATLAB script named myscript.m :
% FILENAME: myscript.m
% Specify pool size.
% Convert the parallel job to a pool job.
parpool('4');
spmd
if labindex == 1
% Lab (rank) #1 broadcasts an integer value to other labs (ranks).
N = labBroadcast(1,int64(1000));
else
% Each lab (rank) receives the broadcast value from lab (rank) #1.
N = labBroadcast(1);
end
% Form a string with host name, total number of labs, lab ID, and broadcast value.
[c name] =system('hostname');
name = name(1:length(name)-1);
fmt = num2str(floor(log10(numlabs))+1);
str = sprintf(['%s:%d:%' fmt 'd:%d '], name,numlabs,labindex,N);
% Apply global concatenate to all str's.
% Store the concatenation of str's in the first dimension (row) and on lab #1.
result = gcat(str,1,1);
if labindex == 1
disp(result)
end
end % spmd
matlabpool close force;
quit;
Also, prepare a job submission, here named myjob.sub. Run with the name of the script:
# FILENAME: myjob.sub
echo "myjob.sub"
module load matlab
unset DISPLAY
# -nodisplay: run MATLAB in text mode; X11 server not needed
# -r: read MATLAB program; use MATLAB JIT Accelerator
matlab -nodisplay -r myscript
Run MATLAB to set the default parallel configuration to your appropriate Profile:
$ matlab -nodisplay
>> defaultParallelConfig('myslurmprofile');
>> quit;
$
Once this job starts, a second job submission is made.
myjob.sub
< M A T L A B (R) >
Copyright 1984-2011 The MathWorks, Inc.
R2011b (7.13.0.564) 64-bit (glnxa64)
August 13, 2011
To get started, type one of these: helpwin, helpdesk, or demo.
For product information, visit www.mathworks.com.
>Starting matlabpool using the 'myslurmprofile' configuration ... connected to 4 labs.
Lab 1:
resource-a006.rcac.purdue.edu:4:1:1000
resource-a007.rcac.purdue.edu:4:2:1000
resource-a008.rcac.purdue.edu:4:3:1000
resource-a009.rcac.purdue.edu:4:4:1000
Sending a stop signal to all the labs ... stopped.
Did not find any pre-existing parallel jobs created by matlabpool.
Output shows the name of one compute node (a006) that processed the job submission file myjob.sub. The job submission scattered four processor cores (four MATLAB labs) among four different compute nodes (a006,a007,a008,a009) that processed the four parallel regions.
To scale up this method to handle a real application, increase the wall time in the submission command to accommodate a longer running job. Secondly, increase the wall time of myslurmprofile by using the Cluster Profile Manager in the Parallel menu to enter a new wall time in the property SubmitArguments.
For more information about parallel jobs:
meep
Link to section 'Description' of 'meep' Description
Meep or MEEP is a free finite-difference time-domain FDTD simulation software package developed at MIT to model electromagnetic systems.
Link to section 'Versions' of 'meep' Versions
- Negishi: 1.20.0
- Anvil: 1.20.0
Link to section 'Module' of 'meep' Module
You can load the modules by:
module load meep
modtree
Link to section 'Description' of 'modtree' Description
ModuleTree or modtree helps users naviagate between different application stacks and sets up a default compiler and mpi environment.
Link to section 'Versions' of 'modtree' Versions
- Bell: deprecated, new
- Scholar: deprecated, recent
- Gilbreth: deprecated, new
- Negishi: cpu
- Anvil: cpu, gpu
- Gautschi: cpu
Link to section 'Module' of 'modtree' Module
You can load the modules by:
module load modtree
monitor
Link to section 'Description' of 'monitor' Description
System resource monitoring tool.
Link to section 'Versions' of 'monitor' Versions
- Anvil: 2.3.1
- Negishi: 2.3.1
- Gautschi: 2.3.1
Link to section 'Module' of 'monitor' Module
You can load the modules by:
module load monitor
mpc
Link to section 'Description' of 'mpc' Description
Gnu Mpc is a C library for the arithmetic of complex numbers with arbitrarily high precision and correct rounding of the result.
Link to section 'Versions' of 'mpc' Versions
- Bell: 1.1.0
- Gilbreth: 1.1.0
- Negishi: 1.1.0
- Anvil: 1.1.0
- Gautschi: 1.3.1
Link to section 'Module' of 'mpc' Module
You can load the modules by:
module load mpc
mpfr
Link to section 'Description' of 'mpfr' Description
The MPFR library is a C library for multiple-precision floating-point computations with correct rounding.
Link to section 'Versions' of 'mpfr' Versions
- Bell: 3.1.6, 4.2.1
- Gilbreth: 3.1.6
- Negishi: 4.0.2
- Anvil: 3.1.6, 4.0.2
Link to section 'Module' of 'mpfr' Module
You can load the modules by:
module load mpfr
mrbayes
Link to section 'Description' of 'mrbayes' Description
MrBayes is a program for Bayesian inference and model choice across a wide range of phylogenetic and evolutionary models. MrBayes uses Markov chain Monte Carlo MCMC methods to estimate the posterior distribution of model parameters.
Link to section 'Versions' of 'mrbayes' Versions
- Anvil: 3.2.7a
- Bell: 3.2.7
- Negishi: 3.2.7
- Scholar: 3.2.7
- Gautschi: 3.2.7
Link to section 'Module' of 'mrbayes' Module
You can load the modules by:
module load mrbayes
mxnet
Link to section 'Description' of 'mxnet' Description
NVIDIA Optimized Deep Learning Framework, powered by Apache MXNet is a deep learning framework designed for both efficiency and flexibility. It allows you to mix the flavors of symbolic programming and imperative programming to maximize efficiency and productivity.
Link to section 'Versions' of 'mxnet' Versions
- Gilbreth: 1.7.0
Link to section 'Module' of 'mxnet' Module
You can load the modules by:
module load mxnet
namd
Link to section 'Description' of 'namd' Description
NAMDis a parallel molecular dynamics code designed for high-performance simulation of large biomolecular systems.
Link to section 'Versions' of 'namd' Versions
- Anvil: 2.14, 3.0
- Bell: 2.15, 3.0
- Gilbreth: 2.13, 3.0
- Negishi: 2.14, 2.15, 3.0
- Scholar: 2.13-multinode, 2.13-singlenode, 3.0-alpha3-singlenode
- Gautschi: 3.0
Link to section 'Module' of 'namd' Module
You can load the modules by:
module load namd
nccl
Link to section 'Description' of 'nccl' Description
Optimized primitives for collective multi-GPU communication.
Link to section 'Versions' of 'nccl' Versions
- Anvil: cuda-11.0_2.11.4, cuda-11.2_2.8.4, cuda-11.4_2.11.4
Link to section 'Module' of 'nccl' Module
You can load the modules by:
module load modtree/gpu
module load nccl
nco
Link to section 'Description' of 'nco' Description
The NCO toolkit manipulates and analyzes data stored in netCDF-accessible formats
Link to section 'Versions' of 'nco' Versions
- Bell: 4.6.7
- Scholar: 5.2.4
- Gilbreth: 4.6.7
- Negishi: 4.9.3, 5.0.1
- Anvil: 4.9.3
- Gautschi: 5.2.4
Link to section 'Module' of 'nco' Module
You can load the modules by:
module load nco
ncview
Link to section 'Description' of 'ncview' Description
Simple viewer for NetCDF files.
Link to section 'Versions' of 'ncview' Versions
- Bell: 2.1.7
- Scholar: 2.1.9
- Gilbreth: 2.1.7
- Negishi: 2.1.9
- Anvil: 2.1.8
- Gautschi: 2.1.9
Link to section 'Module' of 'ncview' Module
You can load the modules by:
module load ncview
netcdf-c
Link to section 'Description' of 'netcdf-c' Description
NetCDF network Common Data Form is a set of software libraries and machine-independent data formats that support the creation, access, and sharing of array-oriented scientific data. This is the C distribution.
Link to section 'Versions' of 'netcdf-c' Versions
- Bell: 4.9.0
- Negishi: 4.9.0
- Anvil: 4.7.4
- Gautschi: 4.9.2
- Scholar: 4.9.2
Link to section 'Module' of 'netcdf-c' Module
You can load the modules by:
module load netcdf-c
netcdf-cxx4
Link to section 'Description' of 'netcdf-cxx4' Description
NetCDF network Common Data Form is a set of software libraries and machine-independent data formats that support the creation, access, and sharing of array-oriented scientific data. This is the C++ distribution.
Link to section 'Versions' of 'netcdf-cxx4' Versions
- Bell: 4.3.0, 4.3.1
- Scholar: 4.3.1
- Gilbreth: 4.3.0, 4.3.1
- Negishi: 4.3.1
- Anvil: 4.3.1
- Gautschi: 4.3.1
Link to section 'Module' of 'netcdf-cxx4' Module
You can load the modules by:
module load netcdf-cxx4
netcdf-fortran
Link to section 'Description' of 'netcdf-fortran' Description
NetCDF network Common Data Form is a set of software libraries and machine-independent data formats that support the creation, access, and sharing of array-oriented scientific data. This is the Fortran distribution.
Link to section 'Versions' of 'netcdf-fortran' Versions
- Bell: 4.4.4, 4.5.3, 4.6.0
- Scholar: 4.6.1
- Gilbreth: 4.4.4, 4.5.2, 4.6.1
- Negishi: 4.6.0
- Anvil: 4.5.3
- Gautschi: 4.6.1
Link to section 'Module' of 'netcdf-fortran' Module
You can load the modules by:
module load netcdf-fortran
netlib-lapack
Link to section 'Description' of 'netlib-lapack' Description
LAPACK version 3.X is a comprehensive FORTRAN library that does linear algebra operations including matrix inversions, least squared solutions to linear sets of equations, eigenvector analysis, singular value decomposition, etc. It is a very comprehensive and reputable package that has found extensive use in the scientific community.
Link to section 'Versions' of 'netlib-lapack' Versions
- Bell: 3.8.0
- Scholar: 3.11.0
- Gilbreth: 3.6.0, 3.8.0
- Negishi: 3.8.0
- Anvil: 3.8.0
- Gautschi: 3.11.0
Link to section 'Module' of 'netlib-lapack' Module
You can load the modules by:
module load netlib-lapack
numactl
Link to section 'Description' of 'numactl' Description
Simple NUMA policy support. It consists of a numactl program to run other programs with a specific NUMA policy and a libnuma shared library ("NUMA API") to set NUMA policy in applications.
Link to section 'Versions' of 'numactl' Versions
- Bell: 2.0.14
- Negishi: 2.0.14
- Anvil: 2.0.14
Link to section 'Module' of 'numactl' Module
You can load the modules by:
module load numactl
Link to section 'Usage' of 'numactl' Usage
numactl [ options ] command {arguments ...} command to run the program
Options:
-H, --hardware,
-m nodes, --membind=nodes, Use the memory on these NUMA nodes
-N nodes, --cpunodebind=nodes, Use the CPUs on these nodes
-C cpus, --physcpubind=cpus, Use these CPUs
nwchem
Link to section 'Description' of 'nwchem' Description
High-performance computational chemistry software
Link to section 'Versions' of 'nwchem' Versions
- Negishi: 7.0.2
- Anvil: 7.0.2
Link to section 'Module' of 'nwchem' Module
You can load the modules by:
module load nwchem
octave
Link to section 'Description' of 'octave' Description
GNU Octave is a high-level language, primarily intended for numerical computations. It provides a convenient command line interface for solving linear and nonlinear problems numerically, and for performing other numerical experiments using a language that is mostly compatible with Matlab. It may also be used as a batch-oriented language.
Link to section 'Versions' of 'octave' Versions
- Bell: 4.4.1
- Negishi: 7.3.0
- Anvil: 6.3.0
Link to section 'Module' of 'octave' Module
You can load the modules by:
module load octave
openblas
Link to section 'Description' of 'openblas' Description
OpenBLAS is an open source implementation of the BLAS API with many hand-crafted optimizations for specific processor types
Link to section 'Versions' of 'openblas' Versions
- Bell: 0.3.8, 0.3.21
- Scholar: 0.3.27
- Gilbreth: 0.2.20, 0.3.7, 0.3.21
- Negishi: 0.3.21
- Anvil: 0.3.17
- Gautschi: 0.3.27
Link to section 'Module' of 'openblas' Module
You can load the modules by:
module load openblas
openfoam
Link to section 'Description' of 'openfoam' Description
OpenFOAM is leading software for computational fluid dynamics (CFD).
Link to section 'Versions' of 'openfoam' Versions
- Bell: 5.x, 9-20211122
- Anvil: 8-20210316
- Negishi: 2312, 9-20211122
- Gautschi:2312
- Scholar:2312
Link to section 'Module' of 'openfoam' Module
You can load the modules by:
module load openmpi/5.0.5
module load openfoam
openjdk
Link to section 'Description' of 'openjdk' Description
The free and open-source java implementation
Link to section 'Versions' of 'openjdk' Versions
- Negishi: 1.8.0_265-b01, 11.0.17_8
- Anvil: 1.8.0_265-b01, 11.0.8_10
Link to section 'Module' of 'openjdk' Module
You can load the modules by:
module load openjdk
panoply
Link to section 'Description' of 'panoply' Description
Panoply is a Java-based cross-platform NetCDF, HDF and GRIB Data Viewer.
Link to section 'Versions' of 'panoply' Versions
- Bell: 4.11.6
- Scholar: 4.11.0
- Gilbreth: 4.11.0
Link to section 'Module' of 'panoply' Module
You can load the modules by:
module load panoply
papi
Link to section 'Description' of 'papi' Description
PAPI provides the tool designer and application engineer with a consistent interface and methodology for use of the performance counter hardware found in most major microprocessors. PAPI enables software engineers to see, in near real time, the relation between software performance and processor events. In addition Component PAPI provides access to a collection of components that expose performance measurement opportunities across the hardware and software stack.
Link to section 'Versions' of 'papi' Versions
- Negishi: 6.0.0.1
- Anvil: 6.0.0.1
Link to section 'Module' of 'papi' Module
You can load the modules by:
module load papi
parafly
Link to section 'Description' of 'parafly' Description
Run UNIX commands in parallel
Link to section 'Versions' of 'parafly' Versions
- Negishi: r2013
- Anvil: r2013
- Gautschi: r2013
Link to section 'Module' of 'parafly' Module
You can load the modules by:
module load parafly
parallel-netcdf
Link to section 'Description' of 'parallel-netcdf' Description
PnetCDF Parallel netCDF is a high-performance parallel I/O library for accessing files in format compatibility with Unidatas NetCDF, specifically the formats of CDF-1, 2, and 5.
Link to section 'Versions' of 'parallel-netcdf' Versions
- Bell: 1.11.2
- Negishi: 1.11.2
- Scholar: 1.12.3
- Anvil: 1.11.2
- Gautschi: 1.12.3
Link to section 'Module' of 'parallel-netcdf' Module
You can load the modules by:
module load parallel-netcdf
paraview
Link to section 'Description' of 'paraview' Description
ParaView is an open-source, multi-platform data analysis and visualization application.
Link to section 'Versions' of 'paraview' Versions
- Bell: 5.6.2
- Anvil: 5.9.1, 5.10.1
- Gilbreth: 5.9.0
- Negishi: 5.12.1, 5.9.1
- Scholar: 5.9.0, 5.13.0
Link to section 'Module' of 'paraview' Module
You can load the modules by:
module load paraview
perl-bioperl
Link to section 'Description' of 'perl-bioperl' Description
BioPerl is the product of a community effort to produce Perl code which is useful in biology. Examples include Sequence objects, Alignment objects and database searching objects. These objects not only do what they are advertised to do in the documentation, but they also interact - Alignment objects are made from the Sequence objects, Sequence objects have access to Annotation and SeqFeature objects and databases, Blast objects can be converted to Alignment objects, and so on. This means that the objects provide a coordinated and extensible framework to do computational biology.
Link to section 'Versions' of 'perl-bioperl' Versions
- Anvil: 1.7.2, 1.7.6
- Bell: 1.7.2
- Negishi: 1.7.2
- Scholar: 1.7.2-pl526
- Gautschi: 1.7.2-pl526
Link to section 'Commands' of 'perl-bioperl' Commands
- bp_aacomp
- bp_bioflat_index
- bp_biogetseq
- bp_chaos_plot
- bp_dbsplit
- bp_extract_feature_seq
- bp_fastam9_to_table
- bp_fetch
- bp_filter_search
- bp_find-blast-matches
- bp_gccalc
- bp_genbank2gff3
- bp_index
- bp_local_taxonomydb_query
- bp_make_mrna_protein
- bp_mask_by_search
- bp_mrtrans
- bp_mutate
- bp_nexus2nh
- bp_nrdb
- bp_oligo_count
- bp_process_gadfly
- bp_process_sgd
- bp_revtrans-motif
- bp_search2alnblocks
- bp_search2gff
- bp_search2table
- bp_search2tribe
- bp_seqconvert
- bp_seqcut
- bp_seq_length
- bp_seqpart
- bp_seqret
- bp_seqretsplit
- bp_split_seq
- bp_sreformat
- bp_taxid4species
- bp_taxonomy2tree
- bp_translate_seq
- bp_tree2pag
- bp_unflatten_seq
Link to section 'Module' of 'perl-bioperl' Module
You can load the modules by:
module load perl-bioperl
petsc
Link to section 'Description' of 'petsc' Description
PETSc is a suite of data structures and routines for the scalable parallel solution of scientific applications modeled by partial differential equations.
Link to section 'Versions' of 'petsc' Versions
- Negishi: 3.17.5, 3.18.3
- Anvil: 3.15.3
Link to section 'Module' of 'petsc' Module
You can load the modules by:
module load petsc
proj
Link to section 'Description' of 'proj' Description
PROJ is a generic coordinate transformation software, that transforms geospatial coordinates from one coordinate reference system CRS to another. This includes cartographic projections as well as geodetic transformations.
Link to section 'Versions' of 'proj' Versions
- Bell: 5.2.0, 8.1.0, 8.2.1
- Gilbreth: 5.2.0, 8.2.1
- Negishi: 5.2.0, 6.2.0, 8.1.0, 8.2.1
- Anvil: 5.2.0, 6.2.0
Link to section 'Module' of 'proj' Module
You can load the modules by:
module load proj
protobuf
Link to section 'Description' of 'protobuf' Description
Protocol Buffers (a.k.a., protobuf) are Google's language-neutral, platform-neutral, extensible mechanism for serializing structured data.
Link to section 'Versions' of 'protobuf' Versions
- Bell: 3.11.4
- Gilbreth: 3.0.2
- Negishi: 3.11.4, 3.18.0
- Anvil: 3.11.4
Link to section 'Module' of 'protobuf' Module
You can load the modules by:
module load protobuf
py-mpi4py
Link to section 'Description' of 'py-mpi4py' Description
mpi4py provides a Python interface to MPI or the Message-Passing Interface. It is useful for parallelizing Python scripts
Link to section 'Versions' of 'py-mpi4py' Versions
- Anvil: 3.0.3
Link to section 'Module' of 'py-mpi4py' Module
You can load the modules by:
module load py-mpi4py
python
Link to section 'Description' of 'python' Description
Native Python 3.9.5 including optimized libraries.
Link to section 'Versions' of 'python' Versions
- Anvil: 3.9.5
- Gautschi: 3.11.9
Link to section 'Module' of 'python' Module
You can load the modules by:
module load python
pytorch
Link to section 'Description' of 'pytorch' Description
PyTorch is a machine learning library with strong support for neural networks and deep learning. PyTorch also has a large user base and software ecosystem.
Link to section 'Versions' of 'pytorch' Versions
- Bell: 1.6.0
- Gilbreth: 1.0.0, 1.4.0, 1.7.0
- Anvil: 1.10.0
- Scholar: 1.5.0, 1.6.0, 1.8.0, 1.9.0, 1.10.0
Link to section 'Module' of 'pytorch' Module
You can load the modules by:
module load learning
module load pytorch
To check the loaded, version, run:
python -c "import torch; print(torch.__version__)"
qt
Link to section 'Description' of 'qt' Description
Qt is a comprehensive cross-platform C++ application framework.
Link to section 'Versions' of 'qt' Versions
- Bell: 5.12.5
- Gilbreth: 5.12.5
- Negishi: 5.15.2
- Anvil: 5.15.2
Link to section 'Module' of 'qt' Module
You can load the modules by:
module load qt
quantum-espresso
Link to section 'Description' of 'quantum-espresso' Description
Quantum ESPRESSO is an integrated suite of Open-Source computer codes for electronic-structure calculations and materials modeling at the nanoscale. It is based on density-functional theory, plane waves, and pseudopotentials.
Link to section 'Versions' of 'quantum-espresso' Versions
- Bell: 6.2.1, 6.3, 6.6
- Scholar: 7.3.1
- Negishi: 7.1
- Anvil: 6.7, 7.2, 7.3
Link to section 'Module' of 'quantum-espresso' Module
You can load the modules by:
module load quantum-espresso
r
Link to section 'Description' of 'r' Description
Linear and nonlinear modelling, statistical tests, time series analysis, classification, clustering, etc. Please consult the R project homepage for further information.
Link to section 'Versions' of 'r' Versions
- Bell: 3.6.3, 4.0.0, 4.1.1, 4.1.2, 4.2.2, 4.3.1, 4.4.1
- Scholar: 4.1.1, 4.4.1
- Gilbreth: 3.6.1, 3.6.3, 4.0.0, 4.1.2, 4.2.2, 4.3.1
- Negishi: 4.1.1, 4.2.2, 4.3.1, 4.4.1
- Anvil: 4.0.5, 4.1.0, 4.1.1, 4.4.1
- Gautschi: 4.1.1, 4.4.1
Link to section 'Module' of 'r' Module
You can load the modules by:
module load r
Link to section 'Setting Up R Preferences with .Rprofile' of 'r' Setting Up R Preferences with .Rprofile
Different clusters have different hardware and softwares. So, if you have access to multiple clusters, you must install your R packages separately for each cluster. Each cluster has multiple versions of R and packages installed with one version of R may not work with another version of R. So, libraries for each R version must be installed in a separate directory. You can define the directory where your R packages will be installed using the environment variable R_LIBS_USER.
For your convenience, a sample .Rprofile example file is provided that can be downloaded to your cluster account and renamed into /.Rprofile (or appended to one) to customize your installation preferences. Detailed instructions:
curl -#LO https://www.rcac.purdue.edu/files/knowledge/run/examples/apps/r/Rprofile_example
mv -ib Rprofile_example ~/.Rprofile
The above installation step needs to be done only once on each of the clusters you have access to. Now load the R module and run R to confirm the unique libPaths:
module load r/4.2.2
R
R> .libPaths()
[1] "/home/zhan4429/R/bell/4.2.2-gcc-9.3.0-xxbnk6s"
[2] "/apps/spack/bell/apps/r/4.2.2-gcc-9.3.0-xxbnk6s/rlib/R/library"
Link to section 'Challenging packages' of 'r' Challenging packages
Below are packages users may have difficulty in installation.
Link to section 'nloptr' of 'r' nloptr
In Bell, the installation may fail due to the default `cmake` version is too old. The solution is easy, users just need to load the newer versions of cmake:
module load cmake/3.20.6
module load r
Rscript -e 'install.packages("nloptr")'
In Brown or other older clusters, because our system's cmake and gcc compilers are old, we may not be able to install the latest version of nloptr. The walkaround is that users can install the older versions of nloptr:
module load r
R
> myrepos = c("https://cran.case.edu")
> install.packages("devtools", repos = myrepos)
> library(devtools)
> install_version("nloptr", version = "> 1.2.2, < 2.0.0", repos = myrepos)
Link to section 'Error: C++17 standard requested but CXX17 is not defined' of 'r' Error: C++17 standard requested but CXX17 is not defined
When users want to install some packages, such as colourvalues, the installation may fail due to Error: C++17 standard requested but CXX17 is not defined. Please follow the below command to fix it:
module load r
module spider gcc
module load gcc/xxx ## the lateste gcc is recommended
mkdir -p ~/.R
echo 'CXX17 = g++ -std=gnu++17 -fPIC' > ~/.R/Makevars
R
> install.packages("xxxx")
Link to section 'RCurl' of 'r' RCurl
Some R packages rely on curl. When you install these packages such as RCurl, you may see such error:checking for curl-config... no Cannot find curl-config To install such packages, you need to load the curl module:
module load curl
module load r
R
> install.packages("RCurl")
Link to section 'raster, stars and sf' of 'r' raster, stars and sf
These R packages have some dependencies. To install them, users will need to load several modules. Note that these modules have multiple versions, and the latest version is recommended. However, the default version may not be the latest version. To check the latest version, please runmodule spider XX.
module spider gdal
module spider geos
module spider proj
module spider sqlite
module load gdal/XXX geos/XXX proj/XXX sqlite/XXX ## XXX is the version to use. The latest version is recommended.
module load r/XXX
R
> install.packages("raster")
install.packages("stars")
install.packages("sf")
Running R jobs
This section illustrates how to submit a small R job to a SLURM queue. The example job computes a Pythagorean triple.
Prepare an R input file with an appropriate filename, here named myjob.R:
# FILENAME: myjob.R
# Compute a Pythagorean triple.
a = 3
b = 4
c = sqrt(a*a + b*b)
c # display result
Prepare a job submission file with an appropriate filename, here named myjob.sub:
#!/bin/bash
# FILENAME: myjob.sub
module load r
# --vanilla:
# --no-save: do not save datasets at the end of an R session
R --vanilla --no-save < myjob.R
For other examples or R jobs:
Installing R packages
Link to section 'Challenges of Managing R Packages in the Cluster Environment' of 'Installing R packages' Challenges of Managing R Packages in the Cluster Environment
- Different clusters have different hardware and softwares. So, if you have access to multiple clusters, you must install your R packages separately for each cluster.
- Each cluster has multiple versions of R and packages installed with one version of R may not work with another version of R. So, libraries for each R version must be installed in a separate directory.
- You can define the directory where your R packages will be installed using the environment variable
R_LIBS_USER. - For your convenience, a sample ~/.Rprofile example file is provided that can be downloaded to your cluster account and renamed into
~/.Rprofile(or appended to one) to customize your installation preferences. Detailed instructions.
Link to section 'Installing Packages' of 'Installing R packages' Installing Packages
-
Step 0: Set up installation preferences.
Follow the steps for setting up your~/.Rprofilepreferences. This step needs to be done only once. If you have created a~/.Rprofilefile previously on a resource, ignore this step. -
Step 1: Check if the package is already installed.
As part of the R installations on community clusters, a lot of R libraries are pre-installed. You can check if your package is already installed by opening an R terminal and entering the commandinstalled.packages(). For example,module load r/4.1.2 Rinstalled.packages()["units",c("Package","Version")] Package Version "units" "0.6-3" quit()If the package you are trying to use is already installed, simply load the library, e.g.,
library('units'). Otherwise, move to the next step to install the package. -
Step 2: Load required dependencies. (if needed)
For simple packages you may not need this step. However, some R packages depend on other libraries. For example, thesfpackage depends ongdalandgeoslibraries. So, you will need to load the corresponding modules before installingsf. Read the documentation for the package to identify which modules should be loaded.module load gdal module load geos -
Step 3: Install the package.
Now install the desired package using the commandinstall.packages('package_name'). R will automatically download the package and all its dependencies from CRAN and install each one. Your terminal will show the build progress and eventually show whether the package was installed successfully or not.Rinstall.packages('sf', repos="https://cran.case.edu/") Installing package into ‘/home/myusername/R/the-resource/4.0.0’ (as ‘lib’ is unspecified) trying URL 'https://cran.case.edu/src/contrib/sf_0.9-7.tar.gz' Content type 'application/x-gzip' length 4203095 bytes (4.0 MB) ================================================== downloaded 4.0 MB ... ... more progress messages ... ... ** testing if installed package can be loaded from final location ** testing if installed package keeps a record of temporary installation path * DONE (sf) The downloaded source packages are in ‘/tmp/RtmpSVAGio/downloaded_packages’ - Step 4: Troubleshooting. (if needed)
If Step 3 ended with an error, you need to investigate why the build failed. Most common reason for build failure is not loading the necessary modules.
Link to section 'Loading Libraries' of 'Installing R packages' Loading Libraries
Once you have packages installed you can load them with the library() function as shown below:
library('packagename')
The package is now installed and loaded and ready to be used in R.
Link to section 'Example: Installing dplyr' of 'Installing R packages' Example: Installing dplyr
The following demonstrates installing the dplyr package assuming the above-mentioned custom ~/.Rprofile is in place (note its effect in the "Installing package into" information message):
module load r
R
install.packages('dplyr', repos="http://ftp.ussg.iu.edu/CRAN/")
Installing package into ‘/home/myusername/R/the-resource/4.0.0’
(as ‘lib’ is unspecified)
...
also installing the dependencies 'crayon', 'utf8', 'bindr', 'cli', 'pillar', 'assertthat', 'bindrcpp', 'glue', 'pkgconfig', 'rlang', 'Rcpp', 'tibble', 'BH', 'plogr'
...
...
...
The downloaded source packages are in
'/tmp/RtmpHMzm9z/downloaded_packages'
library(dplyr)
Attaching package: 'dplyr'
For more information about installing R packages:
Loading Data into R
R is an environment for manipulating data. In order to manipulate data, it must be brought into the R environment. R has a function to read any file that data is stored in. Some of the most common file types like comma-separated variable(CSV) files have functions that come in the basic R packages. Other less common file types require additional packages to be installed. To read data from a CSV file into the R environment, enter the following command in the R prompt:
> read.csv(file = "path/to/data.csv", header = TRUE)
When R reads the file it creates an object that can then become the target of other functions. By default the read.csv() function will give the object the name of the .csv file. To assign a different name to the object created by read.csv enter the following in the R prompt:
> my_variable <- read.csv(file = "path/to/data.csv", header = FALSE)
To display the properties (structure) of loaded data, enter the following:
> str(my_variable)
For more functions and tutorials:
Setting Up R Preferences with .Rprofile
For your convenience, a sample ~/.Rprofile example file is provided that can be downloaded to your cluster account and renamed into ~/.Rprofile (or appended to one). Follow these steps to download our recommended ~/.Rprofile example and copy it into place:
curl -#LO https://www.rcac.purdue.edu/files/knowledge/run/examples/apps/r/Rprofile_example
mv -ib Rprofile_example ~/.Rprofile
The above installation step needs to be done only once on ${resource.name}. Now load the R module and run R:
module load r/4.1.2
R
.libPaths()
[1] "/home/myusername/R/the-resource/4.1.2-gcc-6.3.0-ymdumss"
[2] "/apps/spack/the-resource/apps/r/4.1.2-gcc-6.3.0-ymdumss/rlib/R/library"
.libPaths() should output something similar to above if it is set up correctly.
You are now ready to install R packages into the dedicated directory /home/myusername/R/the-resource/4.1.2-gcc-6.3.0-ymdumss.
rocm
Link to section 'Description' of 'rocm' Description
ROCm Application for Reporting System Info
Link to section 'Versions' of 'rocm' Versions
- Bell: 5.2.0
- Negishi: 6.0.0, 6.1.2
Link to section 'Module' of 'rocm' Module
You can load the modules by:
module load rocm
rstudio
Link to section 'Description' of 'rstudio' Description
This package installs Rstudio desktop from pre-compiled binaries available in the Rstudio website. The installer assumes that you are running on CentOS7/Redhat7/Fedora19. Please fix the download URL for other systems.
Link to section 'Versions' of 'rstudio' Versions
- Bell: 1.3.1073, 1.3.959, 2021.09, 2022.07, 2023.06, 2023.12
- Scholar: 2024.09
- Gilbreth: 1.2.1335, 1.3.959, 2021.09, 2022.07, 2023.06
- Negishi: 2023.06, 2024.04.2
- Anvil: 2021.09.0, 2024.04.2
- Gautschi: 2024.09
Link to section 'Module' of 'rstudio' Module
You can load the modules by:
module load rstudio
samtools
Link to section 'Description' of 'samtools' Description
SAM Tools provide various utilities for manipulating alignments in the SAM format, including sorting, merging, indexing and generating alignments in a per-position format
Link to section 'Versions' of 'samtools' Versions
- Anvil: 1.12, 1.15, 1.16, 1.17, 1.9
- Bell: 1.15, 1.16, 1.17, 1.9
- Negishi: 1.15, 1.16, 1.17, 1.9
- Scholar: 1.15, 1.16, 1.17, 1.9
- Gautschi: 1.9, 1.15, 1.16, 1.17
Link to section 'Commands' of 'samtools' Commands
- ace2sam
- blast2sam.pl
- bowtie2sam.pl
- export2sam.pl
- fasta-sanitize.pl
- interpolate_sam.pl
- maq2sam-long
- maq2sam-short
- md5fa
- md5sum-lite
- novo2sam.pl
- plot-ampliconstats
- plot-bamstats
- psl2sam.pl
- sam2vcf.pl
- samtools
- samtools.pl
- seq_cache_populate.pl
- soap2sam.pl
- wgsim
- wgsim_eval.pl
- zoom2sam.pl
Link to section 'Module' of 'samtools' Module
You can load the modules by:
module load samtools
Link to section 'Example job' of 'samtools' Example job
To run Samtools on our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 1
#SBATCH --job-name=samtools
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load samtools
samtools sort my.sam > my_sorted.bam
samtools index my_sorted.bam
sentaurus
Link to section 'Description' of 'sentaurus' Description
Sentaurus is a suite of TCAD tools which simulates the fabrication, operation and reliability of semiconductor devices. The Sentaurus simulators use physical models to represent the wafer fabrication steps and device operation, thereby allowing the exploration and optimization of new semiconductor devices.
Link to section 'Versions' of 'sentaurus' Versions
- Bell: 2017.09, 2019.03
- Negishi: 2021.06, 2022.3
Link to section 'Module' of 'sentaurus' Module
You can load the modules by:
module load sentaurus
spark
Link to section 'Description' of 'spark' Description
Apache Spark is a fast and general engine for large-scale data processing.
Link to section 'Versions' of 'spark' Versions
- Bell: 2.4.4
- Gilbreth: 2.4.4
- Negishi: 3.1.1
- Anvil: 3.1.1
Link to section 'Module' of 'spark' Module
You can load the modules by:
module load spark
sqlite
Link to section 'Description' of 'sqlite' Description
SQLite3 is an SQL database engine in a C library. Programs that link the SQLite3 library can have SQL database access without running a separate RDBMS process.
Link to section 'Versions' of 'sqlite' Versions
- Bell: 3.30.1, 3.46.0
- Gilbreth: 3.30.1
- Negishi: 3.39.4
- Anvil: 3.35.5
Link to section 'Module' of 'sqlite' Module
You can load the modules by:
module load sqlite
stata-mp
Link to section 'Description' of 'stata-mp' Description
Stata/MP is the fastest and largest edition of Stata. Stata is a complete, integrated software package that provides all your data science needs—data manipulation, visualization, statistics, and automated reporting.
Link to section 'Versions' of 'stata-mp' Versions
- Bell: 17, 18
- Scholar: 17, 18
Link to section 'Module' of 'stata-mp' Module
You can load the modules by:
module load stata-mp
stata
Link to section 'Description' of 'stata' Description
Stata is a complete, integrated software package that provides all your data science needs—data manipulation, visualization, statistics, and automated reporting.
Link to section 'Versions' of 'stata' Versions
- Bell: 18
- Scholar: 17, 18
- Negishi: 18
- Gautschi: 18
Link to section 'Module' of 'stata' Module
You can load the modules by:
module load stata
subversion
Link to section 'Description' of 'subversion' Description
Apache Subversion - an open source version control system.
Link to section 'Versions' of 'subversion' Versions
- Bell: 1.12.2
Link to section 'Module' of 'subversion' Module
You can load the modules by:
module load subversion
swig
Link to section 'Description' of 'swig' Description
SWIG is an interface compiler that connects programs written in C and C++ with scripting languages such as Perl, Python, Ruby, and Tcl. It works by taking the declarations found in C/C++ header files and using them to generate the wrapper code that scripting languages need to access the underlying C/C++ code. In addition, SWIG provides a variety of customization features that let you tailor the wrapping process to suit your application.
Link to section 'Versions' of 'swig' Versions
- Negishi: 4.0.2
- Anvil: 4.0.2
Link to section 'Module' of 'swig' Module
You can load the modules by:
module load swig
tcl
Link to section 'Description' of 'tcl' Description
Tcl Tool Command Language is a very powerful but easy to learn dynamic programming language, suitable for a very wide range of uses, including web and desktop applications, networking, administration, testing and many more. Open source and business-friendly, Tcl is a mature yet evolving language that is truly cross platform, easily deployed and highly extensible.
Link to section 'Versions' of 'tcl' Versions
- Bell: 8.6.8, 8.6.12
- Gilbreth: 8.6.8
- Negishi: 8.6.11, 8.6.12
- Anvil: 8.6.11
Link to section 'Module' of 'tcl' Module
You can load the modules by:
module load tcl
tensorflow
Link to section 'Description' of 'tensorflow' Description
TensorFlow is an open source software library for numerical computation using data flow graphs. Nodes in the graph represent mathematical operations, while the graph edges represent the multidimensional data arrays (tensors) communicated between them. The flexible architecture allows you to deploy computation to one or more CPUs or GPUs in a desktop, server, or mobile device with a single API.
Link to section 'Versions' of 'tensorflow' Versions
- Bell: 2.3.0
- Anvil: 2.6.0
- Gilbreth: 2.4.0, 1.12.0
- Scholar: 1.15.2, 1.15.4, 1.15.5, 2.1.0, 2.2.0, 2.3.1, 2.5.0, 2.6.0
Link to section 'Module' of 'tensorflow' Module
You can load the modules by:
module load learning
module load tensorflow
To check the loaded version, run:
python -c "import tensorflow as tf; print(tf.__version__)"
texinfo
Link to section 'Description' of 'texinfo' Description
Texinfo is the official documentation format of the GNU project. It was invented by Richard Stallman and Bob Chassell many years ago, loosely based on Brian Reids Scribe and other formatting languages of the time. It is used by many non-GNU projects as well.
Link to section 'Versions' of 'texinfo' Versions
- Bell: 6.7
Link to section 'Module' of 'texinfo' Module
You can load the modules by:
module load texinfo
texlive
Link to section 'Description' of 'texlive' Description
TeX Live is a free software distribution for the TeX typesetting system. Heads up, its is not a reproducible installation. At any point only the most recent version can be installed. Older versions are included for backward compatibility, i.e., if you have that version already installed.
Link to section 'Versions' of 'texlive' Versions
- Bell: 20200406, 20220321
- Scholar: 20220321
- Gilbreth: 20200406
- Negishi: 20220321
- Anvil: 20200406
- Gautschi: 20220321
Link to section 'Module' of 'texlive' Module
You can load the modules by:
module load texlive
tflearn
Link to section 'Description' of 'tflearn' Description
TFlearn is a modular and transparent deep learning library built on top of Tensorflow. It was designed to provide a higher-level API to TensorFlow in order to facilitate and speed-up experimentations, while remaining fully transparent and compatible with it.
Link to section 'Versions' of 'tflearn' Versions
- Gilbreth: 0.3.2
Link to section 'Module' of 'tflearn' Module
You can load the modules by:
module load learning
module load tflearn
theano
Link to section 'Description' of 'theano' Description
Theano is a Python library that allows you to define, optimize, and evaluate mathematical expressions involving multi-dimensional arrays efficiently. Theano is most commonly used to perform Deep Learning and has excellent GPU support and integration through PyCUDA.
Link to section 'Versions' of 'theano' Versions
- Bell: 1.0.5
- Gilbreth: 1.0.3, 1.0.5
Link to section 'Module' of 'theano' Module
You can load the modules by:
module load learning
module load theano
thermocalc
Link to section 'Description' of 'thermocalc' Description
Thermo-Calc allows you to calculate the state for a given thermodynamic system to obtain insight.
Link to section 'Versions' of 'thermocalc' Versions
- Bell: 2019b, 2020a, 2021b
Link to section 'Module' of 'thermocalc' Module
You can load the modules by:
module load thermocalc
tk
Link to section 'Description' of 'tk' Description
Tk is a graphical user interface toolkit that takes developing desktop applications to a higher level than conventional approaches. Tk is the standard GUI not only for Tcl, but for many other dynamic languages, and can produce rich, native applications that run unchanged across Windows, Mac OS X, Linux and more.
Link to section 'Versions' of 'tk' Versions
- Bell: 8.6.8, 8.6.11
- Gilbreth: 8.6.8
- Negishi: 8.6.11
- Anvil: 8.6.11
- Gautschi: 8.6.11
Link to section 'Module' of 'tk' Module
You can load the modules by:
module load tk
tophat
Link to section 'Description' of 'tophat' Description
Spliced read mapper for RNA-Seq.
Link to section 'Versions' of 'tophat' Versions
- Anvil: 2.1.1, 2.1.2
- Bell: 2.1.1
- Negishi: 2.1.1
- Scholar: 2.1.1-py27
- Gautschi: 2.1.1-py27
Link to section 'Commands' of 'tophat' Commands
- bam2fastx
- bam_merge
- bed_to_juncs
- contig_to_chr_coords
- fix_map_ordering
- gtf_juncs
- gtf_to_fasta
- juncs_db
- long_spanning_reads
- map2gtf
- prep_reads
- sam_juncs
- samtools_0.1.18
- segment_juncs
- sra_to_solid
- tophat
- tophat2
- tophat-fusion-post
- tophat_reports
Link to section 'Module' of 'tophat' Module
You can load the modules by:
module load tophat
Link to section 'Example job' of 'tophat' Example job
To run TopHat on our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 1
#SBATCH --job-name=tophat
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load tophat
tophat -r 20 test_ref reads_1.fq reads_2.fq
trimmomatic
Link to section 'Description' of 'trimmomatic' Description
A flexible read trimming tool for Illumina NGS data.
Link to section 'Versions' of 'trimmomatic' Versions
- Anvil: 0.39
- Bell: 0.39
- Negishi: 0.39
- Scholar: 0.39
- Gautschi: 0.39
Link to section 'Commands' of 'trimmomatic' Commands
- trimmomatic
Link to section 'Module' of 'trimmomatic' Module
You can load the modules by:
module load trimmomatic
Link to section 'Example job' of 'trimmomatic' Example job
To run Trimmomatic on our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 8
#SBATCH --job-name=trimmomatic
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load trimmomatic
trimmomatic PE -threads 8 \
input_forward.fq.gz input_reverse.fq.gz \
output_forward_paired.fq.gz output_forward_unpaired.fq.gz \
output_reverse_paired.fq.gz output_reverse_unpaired.fq.gz \
ILLUMINACLIP:TruSeq3-PE.fa:2:30:10:2:True LEADING:3 TRAILING:3 MINLEN:36
ucx
Link to section 'Description' of 'ucx' Description
a communication library implementing high-performance messaging for MPI/PGAS frameworks
Link to section 'Versions' of 'ucx' Versions
- Anvil: 1.11.2
- Gilbreth: 1.13.0
Link to section 'Module' of 'ucx' Module
You can load the modules by:
module load ucx
udunits
Link to section 'Description' of 'udunits' Description
Automated units conversion
Link to section 'Versions' of 'udunits' Versions
- Negishi: 2.2.28
- Anvil: 2.2.28
Link to section 'Module' of 'udunits' Module
You can load the modules by:
module load udunits
udunits2
Link to section 'Description' of 'udunits2' Description
Automated units conversion
Link to section 'Versions' of 'udunits2' Versions
- Bell: 2.2.24
- Gilbreth: 2.2.24
Link to section 'Module' of 'udunits2' Module
You can load the modules by:
module load udunits2
valgrind
Link to section 'Description' of 'valgrind' Description
An instrumentation framework for building dynamic analysis.
Link to section 'Versions' of 'valgrind' Versions
- Bell: 3.15.0
- Scholar: 3.22.0
- Gilbreth: 3.13.0
- Negishi: 3.19.0
- Anvil: 3.15.0
Link to section 'Module' of 'valgrind' Module
You can load the modules by:
module load valgrind
vasp
Link to section 'Description' of 'vasp' Description
The Vienna Ab initio Simulation Package VASP is a computer program for atomic scale materials modelling, e.g. electronic structure calculations and quantum-mechanical molecular dynamics, from first principles.
Link to section 'Versions' of 'vasp' Versions
- Anvil: 5.4.4.pl2-vtst, 5.4.4.pl2, 6.3.0-sol, 6.3.0-vtst, 6.3.0-wannier90, 6.3.0
- Gilbreth: 6.4.1
- Negishi: 5.4.4.pl2, 6.4.1_vtst, 6.4.1
Link to section 'Module' of 'vasp' Module
You can load the modules by:
module load vasp
vcftools
Link to section 'Description' of 'vcftools' Description
VCFtools is a program package designed for working with VCF files, such as those generated by the 1000 Genomes Project. The aim of VCFtools is to provide easily accessible methods for working with complex genetic variation data in the form of VCF files.
Link to section 'Versions' of 'vcftools' Versions
- Anvil: 0.1.14, 0.1.16
- Bell: 0.1.16
- Negishi: 0.1.16
- Scholar: 0.1.16
- Gautschi: 0.1.16
Link to section 'Commands' of 'vcftools' Commands
- fill-aa
- fill-an-ac
- fill-fs
- fill-ref-md5
- vcf-annotate
- vcf-compare
- vcf-concat
- vcf-consensus
- vcf-contrast
- vcf-convert
- vcf-fix-newlines
- vcf-fix-ploidy
- vcf-indel-stats
- vcf-isec
- vcf-merge
- vcf-phased-join
- vcf-query
- vcf-shuffle-cols
- vcf-sort
- vcf-stats
- vcf-subset
- vcftools
- vcf-to-tab
- vcf-tstv
- vcf-validator
Link to section 'Module' of 'vcftools' Module
You can load the modules by:
module load vcftools
Link to section 'Example job' of 'vcftools' Example job
To run VCFtools on our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 1
#SBATCH --job-name=vcftools
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load vcftools
vcftools --vcf input_data.vcf --chr 1 \
--from-bp 1000000 --to-bp 2000000
vim
Link to section 'Description' of 'vim' Description
Vim is a highly configurable text editor built to enable efficient text editing. It is an improved version of the vi editor distributed with most UNIX systems. Vim is often called a programmers editor, and so useful for programming that many consider it an entire IDE. Its not just for programmers, though. Vim is perfect for all kinds of text editing, from composing email to editing configuration files.
Link to section 'Versions' of 'vim' Versions
- Bell: 8.1.2141
- Gilbreth: 7.4.2367
- Scholar: 8.2
Link to section 'Module' of 'vim' Module
You can load the modules by:
module load vim
visit
Link to section 'Description' of 'visit' Description
VisIt is an Open Source, interactive, scalable, visualization, animation and analysis tool. Description
Link to section 'Versions' of 'visit' Versions
- Anvil: 3.1.4
- Negishi: 3.3.3
Link to section 'Module' of 'visit' Module
You can load the modules by:
module load visit
vlc
Link to section 'Description' of 'vlc' Description
VLC is a free and open source multimedia player for most multimedia formats.
Link to section 'Versions' of 'vlc' Versions
- Bell: 3.0.9.2
- Gilbreth: 3.0.9.2
- Anvil: 3.0.9.2
- Negishi: 3.0.9.2
Link to section 'Module' of 'vlc' Module
You can load the modules by:
module load vlc
vscode
Link to section 'Description' of 'vscode' Description
Visual Studio Code
Link to section 'Versions' of 'vscode' Versions
- Bell: 1.56, 1.59
- Gilbreth: 1.56, 1.59
- Anvil: 1.61.2
- Negishi: 1.79.2
Link to section 'Module' of 'vscode' Module
You can load the modules by:
module load vscode
vtk
Link to section 'Description' of 'vtk' Description
The Visualization Toolkit VTK is an open-source, freely available software system for 3D computer graphics, image processing and visualization.
Link to section 'Versions' of 'vtk' Versions
- Negishi: 9.0.0
- Anvil: 9.0.0
Link to section 'Module' of 'vtk' Module
You can load the modules by:
module load vtk
wannier90
Link to section 'Description' of 'wannier90' Description
Wannier90 is an open-source code released under GPLv2 for generating maximally-localized Wannier functions and using them to compute advanced electronic properties of materials with high efficiency and accuracy.
Link to section 'Versions' of 'wannier90' Versions
- Anvil: 3.1.0
- Bell: 3.1.0
Link to section 'Module' of 'wannier90' Module
You can load the modules by:
module load wannier90
xalt
Link to section 'Versions' of 'xalt' Versions
- Bell: 1.1.2
- Anvil: 2.10.45
- Scholar: 1.1.2, 2.7.1
- Gilbreth: 1.1.2
- Negishi: 3.0.2
Link to section 'Module' of 'xalt' Module
You can load the modules by:
module load xalt
zlib
Link to section 'Description' of 'zlib' Description
A free, general-purpose, legally unencumbered lossless data-compression library.
Link to section 'Versions' of 'zlib' Versions
- Bell: 1.2.11, 1.2.13
- Gilbreth: 1.2.11, 1.2.11-generic
- Negishi: 1.2.13
- Anvil: 1.2.11
Link to section 'Module' of 'zlib' Module
You can load the modules by:
module load zlib
zstd
Link to section 'Description' of 'zstd' Description
Zstandard, or zstd as short version, is a fast lossless compression algorithm, targeting real-time compression scenarios at zlib-level and better compression ratios. It's backed by a very fast entropy stage, provided by Huff0 and FSE library.
Link to section 'Versions' of 'zstd' Versions
- Negishi: 1.5.2
Link to section 'Module' of 'zstd' Module
You can load the modules by:
module load zstd
nextflow
Link to section 'Description' of 'nextflow' Description
Nextflow is a bioinformatics workflow manager that enables the development of portable and reproducible workflows. It supports deploying workflows on a variety of execution platforms including local, HPC schedulers, AWS Batch, Google Cloud Life Sciences, and Kubernetes. Additionally, it provides support for manage your workflow dependencies through built-in support for Conda, Spack, Docker, Podman, Singularity, Modules, and more.
Link to section 'Versions' of 'nextflow' Versions
- Negishi: 21.10.0, 22.10.1, 23.04.1
- Anvil: 21.10.0, 22.10.1
- Bell: 21.10.0, 22.10.4
- Scholar: 21.10.0
- Gautschi: 21.10.0
Link to section 'Module' of 'nextflow' Module
You can load the modules by:
module load nextflow
Note: Docker is not available on Purdue clusters, so use "-profile singularity", environment modules, or conda for running NextFlow pipelines.
Running Nextflow can be computing or memory intensive. Please do not run it on login nodes, as this might affect other users sharing the same login node with you.
Link to section 'Wrap nextflow into slurm jobscript' of 'nextflow' Wrap nextflow into slurm jobscript
The easiest method to use nextflow on clusters is to place the nextflow run command into a batch script and submitting it to Slurm with sbatch. The manager process will run on the allocated compute node, and all tasks are configured to use the local executor.
#!/bin/bash
#SBATCH -A myQueue
#SBATCH -t 1:00:00
#SBATCH -N 1
#SBATCH -n 12
#SBATCH --job-name=nextflow
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load nextflow
nextflow run main.nf -profile singularity
Link to section 'Nextflow submits tasks as slurm jobs' of 'nextflow' Nextflow submits tasks as slurm jobs
Nextflow can also submit its tasks to Slurm instead of running them on the local host. Place the following file named nextflow.config in your Nextflow working directory:
process {
executor = 'slurm'
queueSize = 50
pollInterval = '1 min'
queueStatInterval = '5 min'
submitRateLimit = '10 sec'
}
Please do not change the above default configuration. Nextflow workflow manager process can generate a disruptive amount of communication requests to Slurm and the configuration file is used to reduce the frequency of those requests.
Link to section 'clusterOptions' of 'nextflow' clusterOptions
Inside the individual process definitions in your scripts, you will need to specify the clusterOptions variable to provide your queue and computing resources appropriate for that task. This can be done by adding something in the pattern of clusterOptions='-A standby -N1 -n1 -c12 -t 1:00:00' to the top of your task process blocks.
Below is a simple example to run Fastqc:
nextflow.enable.dsl=2
process FASTQC {
clusterOptions='-A standby -N1 -n1 -c4 -t 00:30:00'
input:
path reads
script:
"""
mkdir -p fastqc_out
module load biocontainers fastqc
fastqc -o fastqc_out ${reads}
"""
}
reads_ch = Channel.fromPath( 'reads/fastq/*.fastq.gz' )
workflow {
FASTQC(reads_ch)
}
Using clusterOptions='-A standby -N1 -n1 -c4 -t 00:30:00' , each nextflow task will be submitted to standby queue requesting 4 cores and 30 mins walltime.
grace
Link to section 'Description' of 'grace' Description
Grace is a WYSIWYG 2D plotting tool for the X Window System and M*tif.
Link to section 'Versions' of 'grace' Versions
-
Negishi: 5.1.25
Link to section 'Module' of 'grace' Module
You can load the modules by:
module load grace
imagemagick
Link to section 'Description' of 'imagemagick' Description
ImageMagick is a software suite to create, edit, compose, or convert bitmap images.
Link to section 'Versions' of 'imagemagick' Versions
- Bell: 7.0.8-7
- Negishi: 7.0.8-7
Link to section 'Module' of 'imagemagick' Module
You can load the modules by:
module load imagemagick
xcb-util-image
Link to section 'Description' of 'xcb-util-image' Description
The XCB util modules provides a number of libraries which sit on top of libxcb, the core X protocol library, and some of the extension libraries. These experimental libraries provide convenience functions and interfaces which make the raw X protocol more usable. Some of the libraries also provide client-side code which is not strictly part of the X protocol but which have traditionally been provided by Xlib.
Link to section 'Versions' of 'xcb-util-image' Versions
- Negishi: 0.4.0
- Anvil: 0.4.0
- Gautschi: 0.4.1
- Scholar: 0.4.1
Link to section 'Module' of 'xcb-util-image' Module
You can load the modules by:
module load xcb-util-image
xcb-util-keysyms
Link to section 'Description' of 'xcb-util-keysyms' Description
The XCB util modules provides a number of libraries which sit on top of libxcb, the core X protocol library, and some of the extension libraries. These experimental libraries provide convenience functions and interfaces which make the raw X protocol more usable. Some of the libraries also provide client-side code which is not strictly part of the X protocol but which have traditionally been provided by Xlib.
Link to section 'Versions' of 'xcb-util-keysyms' Versions
- Negishi: 0.4.0
- Anvil: 0.4.0
- Gautschi: 0.4.1
- Scholar: 0.4.1
Link to section 'Module' of 'xcb-util-keysyms' Module
You can load the modules by:
module load xcb-util-keysyms
xcb-util-renderutil
Link to section 'Description' of 'xcb-util-renderutil' Description
The XCB util modules provides a number of libraries which sit on top of libxcb, the core X protocol library, and some of the extension libraries. These experimental libraries provide convenience functions and interfaces which make the raw X protocol more usable. Some of the libraries also provide client-side code which is not strictly part of the X protocol but which have traditionally been provided by Xlib.
Link to section 'Versions' of 'xcb-util-renderutil' Versions
- Negishi: 0.3.9
- Anvil: 0.3.9
- Gautschi: 0.3.10
- Scholar: 0.3.10
Link to section 'Module' of 'xcb-util-renderutil' Module
You can load the modules by:
module load xcb-util-renderutil
xcb-util-wm
Link to section 'Description' of 'xcb-util-wm' Description
The XCB util modules provides a number of libraries which sit on top of libxcb, the core X protocol library, and some of the extension libraries. These experimental libraries provide convenience functions and interfaces which make the raw X protocol more usable. Some of the libraries also provide client-side code which is not strictly part of the X protocol but which have traditionally been provided by Xlib.
Link to section 'Versions' of 'xcb-util-wm' Versions
- Negishi: 0.4.1
- Anvil: 0.4.1
- Gautschi: 0.4.2
- Scholar: 0.4.2
Link to section 'Module' of 'xcb-util-wm' Module
You can load the modules by:
module load xcb-util-wm
libxscrnsaver
Link to section 'Description' of 'libxscrnsaver' Description
XScreenSaver - X11 Screen Saver extension client library
Link to section 'Versions' of 'libxscrnsaver' Versions
- Negishi: 1.2.2
- Anvil: 1.2.2
- Gautschi: 1.2.2
- Scholar: 1.2.2
Link to section 'Module' of 'libxscrnsaver' Module
You can load the modules by:
module load libxscrnsaver
libxslt
Link to section 'Description' of 'libxslt' Description
Libxslt is the XSLT C library developed for the GNOME project. XSLT itself is a an XML language to define transformation for XML. Libxslt is based on libxml2 the XML C library developed for the GNOME project. It also implements most of the EXSLT set of processor-portable extensions functions and some of Saxons evaluate and expressions extensions.
Link to section 'Versions' of 'libxslt' Versions
- Negishi: 1.1.33
- Anvil: 1.1.33
- Gautschi: 1.1.33
- Scholar: 1.1.33
Link to section 'Module' of 'libxslt' Module
You can load the modules by:
module load libxslt
mesa-glu
Link to section 'Description' of 'mesa-glu' Description
This package provides the Mesa OpenGL Utility library.
Link to section 'Versions' of 'mesa-glu' Versions
- Negishi: 9.0.2
- Anvil: 9.0.2
- Gautschi: 9.0.2
- Scholar: 9.0.2
Link to section 'Module' of 'mesa-glu' Module
You can load the modules by:
module load mesa-glu
motif
Link to section 'Description' of 'motif' Description
Motif - Graphical user interface GUI specification and the widget toolkit
Link to section 'Versions' of 'motif' Versions
- Negishi: 2.3.8
- Gautschi: 2.3.8
- Scholar: 2.3.8
Link to section 'Module' of 'motif' Module
You can load the modules by:
module load motif
Singularity
Note: Singularity was originally a project out of Lawrence Berkeley National Laboratory. It has now been spun off into a distinct offering under a new corporate entity under the name Sylabs Inc. This guide pertains to the open source community edition, SingularityCE.
Link to section 'What is Singularity?' of 'Singularity' What is Singularity?
Singularity is a new feature of the Community Clusters allowing the portability and reproducibility of operating system and application environments through the use of Linux containers. It gives users complete control over their environment.
Singularity is like Docker but tuned explicitly for HPC clusters. More information is available from the project’s website.
Link to section 'Features' of 'Singularity' Features
- Run the latest applications on an Ubuntu or Centos userland
- Gain access to the latest developer tools
- Launch MPI programs easily
- Much more
Singularity’s user guide is available at: sylabs.io/guides/3.8/user-guide
Link to section 'Example' of 'Singularity' Example
Here is an example using an Ubuntu 16.04 image on ${resource.name}:
singularity exec /depot/itap/singularity/ubuntu1604.img cat /etc/lsb-release
DISTRIB_ID=Ubuntu
DISTRIB_RELEASE=16.04
DISTRIB_CODENAME=xenial
DISTRIB_DESCRIPTION="Ubuntu 16.04 LTS"
Here is another example using a Centos 7 image:
singularity exec /depot/itap/singularity/centos7.img cat /etc/redhat-release
CentOS Linux release 7.2.1511 (Core)
Link to section 'Purdue Cluster Specific Notes' of 'Singularity' Purdue Cluster Specific Notes
All service providers will integrate Singularity slightly differently depending on site. The largest customization will be which default files are inserted into your images so that routine services will work.
Services we configure for your images include DNS settings and account information. File systems we overlay into your images are your home directory, scratch, Data Depot, and application file systems.
Here is a list of paths:
- /etc/resolv.conf
- /etc/hosts
- /home/$USER
- /apps
- /scratch
- /depot
This means that within the container environment these paths will be present and the same as outside the container. The /apps, /scratch, and /depot directories will need to exist inside your container to work properly.
Link to section 'Creating Singularity Images' of 'Singularity' Creating Singularity Images
Due to how singularity containers work, you must have root privileges to build an image. Once you have a singularity container image built on your own system, you can copy the image file up to the cluster (you do not need root privileges to run the container).
You can find information and documentation for how to install and use singularity on your system:
We have version 3.8.0-1.el7 on the cluster. You will most likely not be able to run any container built with any singularity past that version. So be sure to follow the installation guide for version 3.8 on your system.
singularity --version
singularity version 3.8.0-1.el7
Everything you need on how to build a container is available from their user-guide. Below are merely some quick tips for getting your own containers built for ${resource.name}.
You can use a Definition File to both build your container and share its specification with collaborators (for the sake of reproducibility). Here is a simplistic example of such a file:
# FILENAME: Buildfile
Bootstrap: docker
From: ubuntu:18.04
%post
apt-get update && apt-get upgrade -y
mkdir /apps /depot /scratch
To build the image itself:
sudo singularity build ubuntu-18.04.sif Buildfile
The challenge with this approach however is that it must start from scratch if you decide to change something. In order to create a container image iteratively and interactively, you can use the --sandbox option.
sudo singularity build --sandbox ubuntu-18.04 docker://ubuntu:18.04
This will not create a flat image file but a directory tree (i.e., a folder), the contents of which are the container's filesystem. In order to get a shell inside the container that allows you to modify it, user the --writable option.
sudo singularity shell --writable ubuntu-18.04
Singularity: Invoking an interactive shell within container...
Singularity ubuntu-18.04.sandbox:~>
You can then proceed to install any libraries, software, etc. within the container. Then to create the final image file, exit the shell and call the build command once more on the sandbox.
sudo singularity build ubuntu-18.04.sif ubuntu-18.04
Finally, copy the new image to ${resource.name} and run it.
Apptainer
Note: Apptainer was formerly known as Singularity and is now a part of the Linux Foundation. When migrating from Singularity see the user compatibility documentation.
Link to section 'What is Apptainer?' of 'Apptainer' What is Apptainer?
Apptainer is an open-source container platform designed to be simple, fast, and secure. It allows the portability and reproducibility of operating systems and application environments through the use of Linux containers. It gives users complete control over their environment.
Apptainer is like Docker but tuned explicitly for HPC clusters. More information is available on the project’s website.
Link to section 'Features' of 'Apptainer' Features
- Run the latest applications on an Ubuntu or Centos userland
- Gain access to the latest developer tools
- Launch MPI programs easily
- Much more
Apptainer’s user guide is available at: apptainer.org/docs/user/main/introduction.html
Link to section 'Example' of 'Apptainer' Example
Here is an example using an Ubuntu 16.04 image on ${resource.name}:
apptainer exec /depot/itap/singularity/ubuntu1604.img cat /etc/lsb-release
DISTRIB_ID=Ubuntu
DISTRIB_RELEASE=16.04
DISTRIB_CODENAME=xenial
DISTRIB_DESCRIPTION="Ubuntu 16.04 LTS"
Here is another example using a Centos 7 image:
apptainer exec /depot/itap/singularity/centos7.img cat /etc/redhat-release
CentOS Linux release 7.3.1611 (Core)
Link to section 'Purdue Cluster Specific Notes' of 'Apptainer' Purdue Cluster Specific Notes
All service providers will integrate Apptainer slightly differently depending on site. The largest customization will be which default files are inserted into your images so that routine services will work.
Services we configure for your images include DNS settings and account information. File systems we overlay into your images are your home directory, scratch, Data Depot, and application file systems.
Here is a list of paths:
- /etc/resolv.conf
- /etc/hosts
- /home/$USER
- /apps
- /scratch
- /depot
This means that within the container environment these paths will be present and the same as outside the container. The /apps, /scratch, and /depot directories will need to exist inside your container to work properly.
Link to section 'Creating Apptainer Images' of 'Apptainer' Creating Apptainer Images
You can build on your system or straight on the cluster (you do not need root privileges to build or run the container).
You can find information and documentation for how to install and use Apptainer on your system:
We have version 1.1.6 (or newer) on the cluster. Please note that installed versions may change throughout cluster life time, so when in doubt, please check exact version with a --version command line flag:
apptainer --version
apptainer version 1.1.6-1
Everything you need on how to build a container is available from their user guide. Below are merely some quick tips for getting your own containers built for ${resource.name}.
You can use a Definition File to both build your container and share its specification with collaborators (for the sake of reproducibility). Here is a simplistic example of such a file:
# FILENAME: Buildfile
Bootstrap: docker
From: ubuntu:18.04
%post
apt-get update && apt-get upgrade -y
mkdir /apps /depot /scratch
To build the image itself:
apptainer build ubuntu-18.04.sif Buildfile
The challenge with this approach however is that it must start from scratch if you decide to change something. In order to create a container image iteratively and interactively, you can use the --sandbox option.
apptainer build --sandbox ubuntu-18.04 docker://ubuntu:18.04
This will not create a flat image file but a directory tree (i.e., a folder), the contents of which are the container's filesystem. In order to get a shell inside the container that allows you to modify it, user the --writable option.
apptainer shell --writable ubuntu-18.04
Apptainer>
You can then proceed to install any libraries, software, etc. within the container. Then to create the final image file, exit the shell and call the build command once more on the sandbox.
apptainer build ubuntu-18.04.sif ubuntu-18.04
Finally, copy the new image to ${resource.name} and run it.
tecplot
Link to section 'Description' of 'tecplot' Description
Tecplot 360 is a Computational Fluid Dynamics (CFD) and numerical simulation software package used in post-processing simulation results. It is also used in chemistry applications to visualize molecule structure by post-processing charge density data.
Link to section 'Versions' of 'tecplot' Versions
- Bell: 360-2017-R3, 360-2021-R1
- Scholar: 360-2017-R3
- Gilbreth: 360-2017-R3
- Negishi: 360-2022-R2, 360-2024-R1
- Gautschi: 360-2024-R1
Link to section 'Module' of 'tecplot' Module
You can load the modules by:
module load tecplot
ncl
Link to section 'Description' of 'ncl' Description
NCL is an interpreted language designed specifically for scientific data analysis and visualization. Supports NetCDF 3/4, GRIB 1/2, HDF 4/5, HDF-EOD 2/5, shapefile, ASCII, binary. Numerous analysis functions are built-in.
Link to section 'Versions' of 'ncl' Versions
- Bell: 6.4.0
- Scholar: 6.6.2
- Gilbreth: 6.4.0
- Negishi: 6.6.2
- Anvil: 6.4.0
- Gautschi: 6.6.2
Link to section 'Module' of 'ncl' Module
You can load the modules by:
module load ncl
vmd
Link to section 'Description' of 'vmd' Description
VMD is a molecular visualization program for displaying, animating, and analyzing large biomolecular systems using 3-D graphics and built-in scripting.
Link to section 'Versions' of 'vmd' Versions
- Bell: 1.9.3
- Gilbreth: 1.9.3
- Anvil: 1.9.3
Link to section 'Module' of 'vmd' Module
You can load the modules by:
module load vmd
nf-core
Link to section 'Description' of 'nf-core' Description
A community effort to collect a curated set of analysis pipelines built using Nextflow and tools to run the pipelines.
Home page: https://nf-co.re
Link to section 'Versions' of 'nf-core' Versions
- Anvil: 2.7.2, 2.8
- Bell: 2.8
- Negishi: 2.7.2, 2.8, 2.11.1
Link to section 'Commands' of 'nf-core' Commands
___ __ __ __ ___ /,-._.--~\
|\ | |__ __ / ` / \ |__) |__ } {
| \| | \__, \__/ | \ |___ \`-._,-`-,
`._,._,'
nf-core/tools version 2.8 - https://nf-co.re
Usage: nf-core [OPTIONS] COMMAND [ARGS]...
nf-core/tools provides a set of helper tools for use with nf-core Nextflow pipelines.
It is designed for both end-users running pipelines and also developers creating new pipelines.
╭─ Options ────────────────────────────────────────────────────────────────────────────────────────╮
│ --version Show the version and exit. │
│ --verbose -v Print verbose output to the console. │
│ --hide-progress Don't show progress bars. │
│ --log-file -l Save a verbose log to a file. │
│ --help -h Show this message and exit. │
╰──────────────────────────────────────────────────────────────────────────────────────────────────╯
╭─ Commands for users ─────────────────────────────────────────────────────────────────────────────╮
│ list List available nf-core pipelines with local info. │
│ launch Launch a pipeline using a web GUI or command line prompts. │
│ download Download a pipeline, nf-core/configs and pipeline singularity images. │
│ licences List software licences for a given workflow (DSL1 only). │
╰──────────────────────────────────────────────────────────────────────────────────────────────────╯
╭─ Commands for developers ────────────────────────────────────────────────────────────────────────╮
│ create Create a new pipeline using the nf-core template. │
│ lint Check pipeline code against nf-core guidelines. │
│ modules Commands to manage Nextflow DSL2 modules (tool wrappers). │
│ subworkflows Commands to manage Nextflow DSL2 subworkflows (tool wrappers). │
│ schema Suite of tools for developers to manage pipeline schema. │
│ bump-version Update nf-core pipeline version number. │
│ sync Sync a pipeline TEMPLATE branch with the nf-core template. │
╰──────────────────────────────────────────────────────────────────────────────────────────────────╯
Link to section 'Module' of 'nf-core' Module
You can load the modules by:
module load nf-core
Link to section 'List available pipelines' of 'nf-core' List available pipelines
To check all available pipelines:
$ nf-core list
,--./,-.
___ __ __ __ ___ /,-._.--~\
|\ | |__ __ / ` / \ |__) |__ } {
| \| | \__, \__/ | \ |___ \`-._,-`-,
`._,._,'
nf-core/tools version 2.8 - https://nf-co.re
┏━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━┳━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Pipeline Name ┃ Stars ┃ Latest Release ┃ Released ┃ Last Pulled ┃ Have latest release? ┃
┡━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━╇━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━┩
│ funcscan │ 29 │ 1.1.0 │ 2 days ago │ - │ - │
│ smrnaseq │ 49 │ 2.2.0 │ 4 days ago │ - │ - │
│ rnafusion │ 95 │ 2.3.4 │ 4 days ago │ - │ - │
│ rnaseq │ 604 │ 3.11.2 │ 5 days ago │ 5 days ago │ No (dev - 4b7695a) │
│ demultiplex │ 26 │ 1.2.0 │ 5 days ago │ - │ - │
│ differentialabundan… │ 19 │ 1.2.0 │ 2 weeks ago │ 2 days ago │ Yes (v1.2.0) │
│ mhcquant │ 21 │ 2.4.1 │ 3 weeks ago │ - │ - │
│ viralintegration │ 8 │ 0.1.0 │ 1 months ago │ - │ - │
│ quantms │ 8 │ 1.1.1 │ 1 months ago │ - │ - │
│ viralrecon │ 93 │ 2.6.0 │ 1 months ago │ - │ - │
│ airrflow │ 24 │ 3.0 │ 1 months ago │ - │ - │
│ scrnaseq │ 81 │ 2.2.0 │ 1 months ago │ - │ - │
│ epitopeprediction │ 25 │ 2.2.1 │ 1 months ago │ - │ - │
│ isoseq │ 12 │ 1.1.4 │ 2 months ago │ - │ - │
│ taxprofiler │ 49 │ 1.0.0 │ 2 months ago │ 2 weeks ago │ No (master - c3f1adf) │
│ nanoseq │ 109 │ 3.1.0 │ 2 months ago │ - │ - │
│ cutandrun │ 41 │ 3.1 │ 2 months ago │ - │ - │
│ circdna │ 12 │ 1.0.2 │ 2 months ago │ - │ - │
│ ampliseq │ 111 │ 2.5.0 │ 2 months ago │ - │ - │
│ mag │ 126 │ 2.3.0 │ 2 months ago │ - │ - │
│ nascent │ 8 │ 2.1.1 │ 2 months ago │ - │ - │
│ phyloplace │ 3 │ 1.0.0 │ 2 months ago │ - │ - │
│ proteinfold │ 21 │ 1.0.0 │ 3 months ago │ - │ - │
│ crisprseq │ 8 │ 1.0 │ 3 months ago │ - │ - │
│ hic │ 48 │ 2.0.0 │ 3 months ago │ 2 weeks ago │ Yes (v2.0.0) │
│ sarek │ 235 │ 3.1.2 │ 4 months ago │ - │ - │
│ fetchngs │ 78 │ 1.9 │ 4 months ago │ - │ - │
│ methylseq │ 104 │ 2.3.0 │ 4 months ago │ - │ - │
│ atacseq │ 134 │ 2.0 │ 5 months ago │ - │ - │
│ eager │ 91 │ 2.4.6 │ 5 months ago │ 2 days ago │ Yes (v2.4.6) │
│ coproid │ 7 │ 1.1.1 │ 6 months ago │ - │ - │
│ hgtseq │ 16 │ 1.0.0 │ 6 months ago │ - │ - │
│ hlatyping │ 41 │ 2.0.0 │ 6 months ago │ - │ - │
│ chipseq │ 144 │ 2.0.0 │ 7 months ago │ - │ - │
│ rnavar │ 16 │ 1.0.0 │ 10 months ago │ - │ - │
│ mnaseseq │ 9 │ 1.0.0 │ 11 months ago │ - │ - │
│ hicar │ 3 │ 1.0.0 │ 12 months ago │ - │ - │
│ bamtofastq │ 8 │ 1.2.0 │ 1 years ago │ - │ - │
│ bacass │ 42 │ 2.0.0 │ 2 years ago │ 5 days ago │ Yes (v2.0.0) │
│ bactmap │ 41 │ 1.0.0 │ 2 years ago │ - │ - │
│ metaboigniter │ 10 │ 1.0.1 │ 2 years ago │ - │ - │
│ diaproteomics │ 10 │ 1.2.4 │ 2 years ago │ - │ - │
│ clipseq │ 13 │ 1.0.0 │ 2 years ago │ - │ - │
│ pgdb │ 3 │ 1.0.0 │ 2 years ago │ - │ - │
│ dualrnaseq │ 12 │ 1.0.0 │ 2 years ago │ - │ - │
│ cageseq │ 9 │ 1.0.2 │ 2 years ago │ - │ - │
│ proteomicslfq │ 29 │ 1.0.0 │ 3 years ago │ - │ - │
│ imcyto │ 20 │ 1.0.0 │ 3 years ago │ - │ - │
│ slamseq │ 4 │ 1.0.0 │ 3 years ago │ - │ - │
│ callingcards │ 1 │ dev │ - │ - │ - │
│ circrna │ 27 │ dev │ - │ - │ - │
│ fastquorum │ 8 │ dev │ - │ - │ - │
│ genomeannotator │ 9 │ dev │ - │ - │ - │
│ genomeassembler │ 12 │ dev │ - │ - │ - │
│ gwas │ 12 │ dev │ - │ - │ - │
│ lncpipe │ 25 │ dev │ - │ - │ - │
│ metapep │ 3 │ dev │ - │ - │ - │
│ metatdenovo │ 2 │ dev │ - │ - │ - │
│ nanostring │ 2 │ dev │ - │ - │ - │
│ pangenome │ 23 │ dev │ - │ 2 weeks ago │ No (a_brave_new_world │
│ │ │ │ │ │ - 6aa9b39) │
│ radseq │ 0 │ dev │ - │ - │ - │
│ raredisease │ 37 │ dev │ - │ - │ - │
│ rnadnavar │ 0 │ dev │ - │ - │ - │
│ rnasplice │ 3 │ dev │ - │ - │ - │
│ scflow │ 19 │ dev │ - │ - │ - │
│ spatialtranscriptom… │ 19 │ dev │ - │ - │ - │
│ spinningjenny │ 0 │ dev │ - │ - │ - │
│ variantcatalogue │ 3 │ dev │ - │ - │ - │
└──────────────────────┴───────┴────────────────┴───────────────┴─────────────┴───────────────────────┘
Link to section 'Download pipelines' of 'nf-core' Download pipelines
It is highly recommended to download pipelines to clusters before running them. Using singularity containers to run these pipelines are also recommended. Befor downloading, please set up environment variables NXF_SINGULARITY_CACHEDIR for singularity cache or NXF_CONDA_CACHEDIR for conda cache. Below is an example you can add to .bashrc for bash users.
export NXF_SINGULARITY_CACHEDIR="$SCRATCH/singularity/cache"
export NXF_CONDA_CACHEDIR="$SCRATCH/conda/cache"
Below is the example to download the rnaseq pipeline:
$ nf-core download rnaseq
,--./,-.
___ __ __ __ ___ /,-._.--~\
|\ | |__ __ / ` / \ |__) |__ } {
| \| | \__, \__/ | \ |___ \`-._,-`-,
`._,._,'
nf-core/tools version 2.8 - https://nf-co.re
? Select release / branch: (Use arrow keys)
» 3.11.2 [release]
3.11.1 [release]
3.11.0 [release]
3.10.1 [release]
3.10 [release]
3.9 [release]
3.8.1 [release]
3.8 [release]
3.7 [release]
3.6 [release]
3.5 [release]
3.4 [release]
3.3 [release]
3.2 [release]
3.1 [release]
3.0 [release]
2.0 [release]
1.4.2 [release]
1.4.1 [release]
1.4 [release]
1.3 [release]
1.2 [release]
1.1 [release]
1.0 [release]
Link to section 'Run the pipeline' of 'nf-core' Run the pipeline
If users have downloaded the rnaseq pipeline to a folder called nf-core under $HOME, users can use it to run RNAseq analysis. Here is an example for running RNAseq analysis with human samples.
#!/bin/bash
#SBATCH -A XXXX
#SBATCH --job-name=rnaseq
#SBATCH --output=slurm-%A.%a.out
#SBATCH --error=slurm-%A.%a.err
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=64
#SBATCH --time=24:00:00
#SBATCH --mail-type=all
module load nextflow
nextflow run $HOME/nf-core/nf-core-rnaseq-3.11.2/workflow/ \
--input samplesheet.csv --outdir results \
--genome GRCh37 -profile singularity
picard
Link to section 'Description' of 'picard' Description
Picard is a set of command line tools for manipulating high-throughput sequencing HTS data and formats such as SAM/BAM/CRAM and VCF.
Link to section 'Versions' of 'picard' Versions
- Anvil: 2.25.1, 2.25.7, 2.26.10, 3.1.1
- Bell: 2.25.1, 2.26.10, 3.1.1
- Negishi: 2.25.1, 2.26.10, 3.1.1
- Scholar: 2.25.1, 2.26.10
- Gautschi: 2.25.1, 2.26.10, 3.1.1
Link to section 'Commands' of 'picard' Commands
- picard
Link to section 'Module' of 'picard' Module
You can load the modules by:
module load picard
Link to section 'Example job' of 'picard' Example job
To run picard our our clusters:
#!/bin/bash
#SBATCH -A myallocation # Allocation name
#SBATCH -t 20:00:00
#SBATCH -N 1
#SBATCH -n 24
#SBATCH --job-name=picard
#SBATCH --mail-type=FAIL,BEGIN,END
#SBATCH --error=%x-%J-%u.err
#SBATCH --output=%x-%J-%u.out
module load picard
picard BuildBamIndex -Xmx64g I=19P0126636WES_sorted_md.bam
picard CreateSequenceDictionary -R hg38.fa -O hg38.dict
sratoolkit
Link to section 'Description' of 'sratoolkit' Description
The NCBI SRA Toolkit enables reading dumping of sequencing files from the SRA database and writing loading files into the .sra format.
Link to section 'Versions' of 'sratoolkit' Versions
- Anvil: 2.10.9
Link to section 'Module' of 'sratoolkit' Module
You can load the modules by:
module load sratoolkit
libv8
Link to section 'Description' of 'libv8' Description
Distributes the V8 JavaScript engine in binary and source forms in order to support fast builds of The Ruby Racer
Link to section 'Versions' of 'libv8' Versions
- Bell: 3.14
- Anvil: 6.7.17
Link to section 'Module' of 'libv8' Module
You can load the modules by:
module load libv8
quantumatk
Link to section 'Versions' of 'quantumatk' Versions
- Bell: 2020.09
- Negishi: 2020.09
Link to section 'Module' of 'quantumatk' Module
You can load the modules by:
module load quantumatk
sas
Link to section 'Description' of 'sas' Description
SAS is a commercial integrated system for statistical analysis, data mining, and graphics as well as many enterprise oriented additional features.
Link to section 'Versions' of 'sas' Versions
- Bell: 9.4
- Scholar: 9.4
- Gilbreth: 9.4
Link to section 'Module' of 'sas' Module
You can load the modules by:
module load sas
netcdf
Link to section 'Description' of 'netcdf' Description
NetCDF network Common Data Form is a set of software libraries and machine-independent data formats that support the creation, access, and sharing of array-oriented scientific data. This is the C distribution.
Link to section 'Versions' of 'netcdf' Versions
- Bell: 4.5.0, 4.7.4
- Scholar: 4.9.2
- Gilbreth: 4.5.0, 4.7.0, 4.9.2
Link to section 'Module' of 'netcdf' Module
You can load the modules by:
module load netcdf
or
module load netcdf-c
libxp
Link to section 'Description' of 'libxp' Description
libXp - X Print Client Library.
Link to section 'Versions' of 'libxp' Versions
- Negishi: 1.0.3
- Gautschi: 1.0.3
- Scholar: 1.0.3
Link to section 'Module' of 'libxp' Module
You can load the modules by:
module load libxp
parallel
Link to section 'Description' of 'parallel' Description
GNU parallel is a shell tool for executing jobs in parallel using one or more computers. A job can be a single command or a small script that has to be run for each of the lines in the input.
Link to section 'Versions' of 'parallel' Versions
- Bell: 20220522
- Negishi: 20220522
- Anvil: 20200822
- Gautschi: 20241022
Link to section 'Module' of 'parallel' Module
You can load the modules by:
module load parallel
Link to section 'Syntax' of 'parallel' Syntax
# Read commands to be run in parallel from an input file
parallel [OPTIONS] < CMDFILE
# Read command arguments on the command line
parallel [OPTIONS] COMMAND [ARGUMENTS] ::: ARGLIST
# Read command arguments from an input file
parallel [OPTIONS] COMMAND [ARGUMENTS] :::: ARGFILE
qemu
Link to section 'Description' of 'qemu' Description
QEMU is a generic and open source machine emulator and virtualizer.
Link to section 'Versions' of 'qemu' Versions
- Bell: 2.10.1, 4.1.0
- Gilbreth: 2.10.1
- Negishi: 4.2.1
- Anvil: 4.1.1, 6.2.0
Link to section 'Module' of 'qemu' Module
You can load the modules by:
module load qemu
totalview
Link to section 'Description' of 'totalview' Description
TotalView is a GUI-based source code defect analysis tool that gives you unprecedented control over processes and thread execution and visibility into program state and variables.
Link to section 'Versions' of 'totalview' Versions
- Bell: 2020.2.6, 2021.4.10
- Gilbreth: 2017.0.12, 2018.2.6, 2019.1.4, 2021.4.10
- Negishi: 2021.4.10
- Anvil: 2020.2.6
Link to section 'Module' of 'totalview' Module
You can load the modules by:
module load totalview
keras
Link to section 'Description' of 'keras' Description
Keras is a deep learning API written in Python, running on top of the machine learning platform TensorFlow. It was developed with a focus on enabling fast experimentation.
Link to section 'Versions' of 'keras' Versions
- Bell: 2.4.3
- Gilbreth: 2.2.4, 2.4.3
- Anvil: 2.6.0
Link to section 'Module' of 'keras' Module
You can load the modules by:
module load learning
module load keras
opencv
Link to section 'Description' of 'opencv' Description
OpenCV, Open source Computer Vision, is an open-source BSD-licensed library that includes several hundreds of computer vision algorithms. Written in optimized C/C++, the library can take advantage of multi-core processing.
Link to section 'Versions' of 'opencv' Versions
- Bell: 4.4.0
- Gilbreth: 3.4.3, 4.2.0, 4.5.1
- Anvil: 4.5.3
Link to section 'Module' of 'opencv' Module
You can load the modules by:
module load learning
module load opencv
libpng
Link to section 'Description' of 'libpng' Description
Libpng is the official open-source PNG reference library. It supports almost all PNG features, is extensible, and has been extensively tested for over 29 years.
Link to section 'Versions' of 'libpng' Versions
- Negishi: 1.5.30, 1.6.37
- Anvil: 1.6.37
- Gautschi: 1.6.37
- Scholar:1.6.37
Link to section 'Module' of 'libpng' Module
You can load the modules by:
module load libpng
ghostscript
Link to section 'Description' of 'ghostscript' Description
Ghostscript is an interpreter for the PostScript® language and PDF files. It is available under either the GNU GPL Affero license or licensed for commercial use from Artifex Software, Inc.
Link to section 'Versions' of 'ghostscript' Versions
- Negishi: 9.56.1
- Anvil: 9.56.1
- Gautschi: 9.56.1
Link to section 'Module' of 'ghostscript' Module
You can load the modules by:
module load libpng
cython
Link to section 'Description' of 'cython' Description
Cython is an optimising static compiler for both the Python programming language and the extended Cython programming language (based on Pyrex). It makes writing C extensions for Python as easy as Python itself.
Link to section 'Versions' of 'cython' Versions
- Gautschi: 3.0.10
Link to section 'Module' of 'cython' Module
You can load the modules by:
module load gcc
module load cython
util-linux-uuid
Link to section 'Description' of 'util-linux-uuid' Description
Util-linux is a suite of essential utilities for any Linux system.
Link to section 'Versions' of 'util-linux-uuid' Versions
- Negishi: 2.38.1
- Anvil: 2.38.1
- Gautschi: 2.38.1
- Scholar: 2.38.1
Link to section 'Module' of 'util-linux-uuid' Module
You can load the modules by:
module load util-linux-uuid
pandoc
Link to section 'Description' of 'pandoc' Description
If you need to convert files from one markup format into another, pandoc is your swiss-army knife.
Link to section 'Versions' of 'pandoc' Versions
- Gautschi: 2.19.2
Link to section 'Module' of 'pandoc' Module
You can load the modules by:
module load pandoc
r-devtools
Link to section 'Description' of 'r-devtools' Description
Tools to Make Developing R Packages Easier. Collection of package development tools.
Link to section 'Versions' of 'r-devtools' Versions
- Gautschi: 2.4.5
- Scholar: 2.4.5
Link to section 'Module' of 'r-devtools' Module
You can load the modules by:
module load r-devtools
r-formatr
Link to section 'Description' of 'r-formatr' Description
Format R Code Automatically. Provides a function tidy_source() to format R source code. Spaces and indent will be added to the code automatically, and comments will be preserved under certain conditions, so that R code will be more human-readable and tidy. There is also a Shiny app as a user interface in this package.
Link to section 'Versions' of 'r-formatr' Versions
- Gautschi: 1.14
- Scholar: 1.14
Link to section 'Module' of 'r-formatr' Module
You can load the modules by:
module load r-formatr
r-irkernel
Link to section 'Description' of 'r-irkernel' Description
Native R Kernel for the 'Jupyter Notebook'. The R kernel for the 'Jupyter' environment executes R code which the front-end ('Jupyter Notebook' or other front-ends) submits to the kernel via the network.
Link to section 'Versions' of 'r-irkernel' Versions
- Gautschi: 1.3.2
- Scholar: 1.3.2
Link to section 'Module' of 'r-irkernel' Module
You can load the modules by:
module load r-irkernel
r-plyr
Link to section 'Description' of 'r-plyr' Description
Tools for Splitting, Applying and Combining Data. A set of tools that solves a common set of problems: you need to break a big problem down into manageable pieces, operate on each piece and then put all the pieces back together. For example, you might want to fit a model to each spatial location or time point in your study, summarise data by panels or collapse high-dimensional arrays to simpler summary statistics. The development of 'plyr' has been generously supported by 'Becton Dickinson'.
Link to section 'Versions' of 'r-plyr' Versions
- Gautschi: 1.8.8
- Scholar: 1.8.8
Link to section 'Module' of 'r-plyr' Module
You can load the modules by:
module load r-plyr
r-reshape2
Link to section 'Description' of 'r-reshape2' Description
Flexibly Reshape Data: A Reboot of the Reshape Package. Flexibly restructure and aggregate data using just two functions: melt and dcast (or acast).
Link to section 'Versions' of 'r-reshape2' Versions
- Gautschi: 1.4.4
- Scholar: 1.4.4
Link to section 'Module' of 'r-reshape2' Module
You can load the modules by:
module load r-reshape2
r-shiny
Link to section 'Description' of 'r-shiny' Description
Web Application Framework for R. Makes it incredibly easy to build interactive web applications with R. Automatic "reactive" binding between inputs and outputs and extensive pre-built widgets make it possible to build beautiful, responsive, and powerful applications with minimal effort.
Link to section 'Versions' of 'r-shiny' Versions
- Gautschi: 1.8.1.1
Link to section 'Module' of 'r-shiny' Module
You can load the modules by:
module load r-shiny
duckdb
Link to section 'Description' of 'duckdb' Description
DuckDB is an in-process SQL OLAP database management system. Simple, feature-rich, fast & open source.
Link to section 'Versions' of 'duckdb' Versions
- Gautschi: 1.1.3
- Scholar: 1.0.0
Link to section 'Module' of 'duckdb' Module
You can load the modules by:
module load duckdb
intel-rt
Link to section 'Description' of 'intel-rt' Description
Intel® oneAPI Compiler redistributable libraries.
Link to section 'Versions' of 'intel-rt' Versions
- Gautschi: 2024.1.0
Link to section 'Module' of 'intel-rt' Module
You can load the modules by:
module load intel-rt
oclfpga
Link to section 'Description' of 'oclfpga' Description
Enables FPGA development using the Intel® oneAPI SYCL Compiler.
Link to section 'Versions' of 'oclfpga' Versions
- Gautschi: 2024.1.0
Link to section 'Module' of 'oclfpga' Module
You can load the modules by:
module load oclfpga
cesm
Link to section 'Description' of 'cesm' Description
Quick start guide for running CESM models on Negishi and Bell clusters
Link to section 'Versions' of 'cesm' Versions
- 2.1.*
- 2.2.*
This guide assumes that the user has read relevant CIME basic usage documentation
Link to section 'Negishi' of 'cesm' Negishi
1. Copy template configuration to your user directory. This can either be Depot, or scratch.
cp -r /depot/itap/tratkus/negishi_cesm_2/CESM /path/to/your/scratch
2. Activate a conda environment which contains all of required dependencies to build CESM model
ml conda
conda activate /depot/itap/tratkus/cesm_conda_environment/general
3. Adjust config_machines.xml, config_compilers.xml, and config_batch.xml with your user and queue information for the Negishi blocks within the files.
vim /CESM/cime/config/cesm/machines/config_machines.xml
Placeholder for block example
vim /CESM/cime/config/cesm/machines/config_compilers.xml
Placeholder for block example
vim /CESM/cime/config/cesm/machines/config_batch.xml
Placeholder for block example
4. Create a case
cd /negishi_cesm2/CESM/cime/scripts
./create_newcase --case /casepath/caseName --compset I2000Clm50Sp --res f09_g17 --machine negishi --run-unsupported --compiler intel
5. Setup, build, and run your case
cd /casePath/caseName
./case.setup
./case.build
./case.submit
SLURM partition requirement fix
Add -p flag to config_batch.xml for Negishi block:
And PARTITION variable to config_machines.xml for Negishi block, under intel environmental variables: